22 September 2020: Meta-Analysis
Prevalence and Prevention of Reproducibility Deficiencies in Life Sciences Research: Large-Scale Meta-Analyses
Nadine M. Mansour12ABCDEF, E. Andrew Balas1ACDE*, Frances M. Yang3CDE, Marlo M. Vernon4BCEFDOI: 10.12659/MSM.922016
Med Sci Monit 2020; 26:e922016
Abstract
BACKGROUND: Studies have found that many published life sciences research results are irreproducible. Our goal was to provide comprehensive risk estimates of familiar reproducibility deficiencies to support quality improvement in research.
MATERIAL AND METHODS: Reports included were peer-reviewed, published between 1980 and 2016, and presented frequency data of basic biomedical research deficiencies. Manual and electronic literature searches were performed in seven bibliographic databases. For deficiency concepts with at least four frequency studies and with a sample size of at least 15 units in each, a meta-analysis was performed.
RESULTS: Overall, 68 publications met our inclusion criteria. The study identified several major groups of research quality defects: study design, cell lines, statistical analysis, and reporting. In the study design group of 3 deficiencies, missing power calculation was the most frequent (82.3% [95% Confidence Interval (CI): 69.9–94.6]). Among the 6 cell line deficiencies, mixed contamination was the most frequent (22.4% [95% CI: 10.4–34.3]). Among the 3 statistical analysis deficiencies, the use of chi-square test when expected cells frequency was <5 was the most prevalent (15.7% [95% CI: –3.2–34.7]). In the reporting group of 12 deficiencies, failure to state the number of tails was the most frequent (65% [95% CI: 39.3–90.8]).
CONCLUSIONS: The results of this study could serve as a general reference when consistently measurable sources of deficiencies need to be identified in research quality improvement.
Keywords: Animal Experimentation, Biomedical Research, Cell Line, Research Design, Research Report, Biological Science Disciplines, Reproducibility of Results
Background
Reproducibility is a crucial requirement of scientific validity. Lack of rigor, non-repeatable research, and quality defects are increasingly mentioned concerns. Much preclinical research may be irreproducible, wasting, by one estimate, billions of dollars in research dollars each year [1]. In some specific types of study (e.g., drug target identification and validation), the majority of published preclinical results could not be validated, implying poor quality research and wasted efforts to replicate [2,3]. Others pointed out that the majority of published biomedical research findings may be unreliable due to the use of invalid statistical methods [4]. The high failure rate of clinical trials is partly blamed on promising but unreliable results coming out of preclinical research [5].
Retractions of scientific papers have also increased 15-fold according to Thomson Reuters Web of Science. Between 2000 and 2010, a large percentage (73.5%) of retractions in medicine and science were withdrawn simply for deficiencies [6,7]. Analysis of 423 retracted articles showed that the most common causes of retractions were laboratory deficiencies (55.8%) and analytical deficiencies (18.9%) and other sources of irreproducibility (16.1%). Cell line contamination was a common cause for retraction in the past, whereas analytical deficiencies were found to be increasing in frequency [8].
The old adage “publish or perish” has elevated tension in the current era of limited funding. According to a study by Foster and colleagues, the majority of published biomedical research studies were based on a traditional model – studying existing known relationships in the biochemistry literature – as opposed to innovation – results that introduce novel relationships, as evidenced by scientific prizes [9]. With the increasing pressures of publications and grant attainment for academics globally, it is no wonder that inadvertent or careless deficiencies appear in scientific research. Additionally, one may also raise the question of economic resources and country income level in deficiency frequencies, especially at a time when the National Institutes of Health (NIH) is implementing policies to promote international biomedical research collaboration [10].
Despite the NIH taking notice that basic biomedical research is most susceptible to reproducibility concerns, the significance of quality defects is still underestimated by the research community [11]. Many researchers are in denial that these quality problems either do not exist or at least “not in my lab”. Meanwhile, the number of articles reporting the frequency of various research deficiencies is steadily increasing.
Measurement of defects is integral to improving the quality of research in the life sciences. Identifying measurable defect frequencies can show measurement opportunities to assess progress of quality improvement and also can guide improvement initiatives by identifying the most frequent types of defects in the research enterprise. To address inaccurate perceptions and to orient improvement efforts, there is a need for risk or frequency estimates of deficiencies based on large and diverse deficiency frequency studies of life sciences research. The purpose of developing this series of meta-analyses was to assist basic scientists as readers and producers of research results by not only itemizing deficiencies responsible for non-reproducible results, but also by providing frequency estimates. In this study, measurements of defects were searched based on the availability necessary published data and also based on the repeated NIH calls to enhancing reproducibility and integrity of research. While this series of analyses was to the extent possible comprehensive, it should not be considered all-inclusive, as the list of recognized and measured research quality deficiencies is continuously evolving.
Material and Methods
ELIGIBILITY CRITERIA:
We identified studies that met the following eligibility criteria: (i) provided a quantitative assessment of the frequency of one or more quality defects in life sciences research (i.e., calculated the frequency of specific deficiencies by dividing the total number of studies showing defect with the total number of studies reporting the particular quality aspect); (ii) presented original frequency data about defects (numerator and denominator); (iii) were peer-reviewed scientific articles that at least had an abstract with numeric results and were written in English; (iv) published between 1980 and 2016.
This study focused on preclinical studies that met stated eligibility criteria. Defects of randomized controlled trials are discussed elsewhere and, therefore, were ineligible for inclusion in this study. Quality aspects that did not meet the criteria, including necessary number of independent studies for a meta-analysis, are recognized but could not be included. Ineligibility criteria also included human clinical trials and articles without online access. Due to the goals of this meta-analysis of deficiency frequency publications, all editorials, commentary, letters, surveys and case reports that did not present data on the frequency of defects were excluded. Studies reporting deficiency frequencies in already known to be defective populations were also ineligible (e.g., studies that analyzed deficiency detection in cell lines that were already known to be contaminated).
SEARCH STRATEGY:
Electronic and manual literature searches were performed to identify all eligible quantitative studies. This study applied a comprehensive search strategy that was based on various combinations of terms and it is available together with all data collected through the Augusta University Scholarly Commons. The searches included the following databases: MEDLINE, CINAHL, Google Scholar, ProQuest Nursing and Allied Health Source, and WOS. The literature search strategy was developed using medical subject headings (MeSH) as well as all key terms related to the following 4 research deficiency groups: study design, statistical analysis, reporting, and cell line. Numerous iterations and combinations of search expressions and phrases were used to achieve maximum retrieval. For example, search terms included “statistical analysis”, “methodology”, “statistical method”, “inappropriate design”, “cell line authentication”, and “contamination in cell lines” and others in combination with terms of “deficiency,” “defect,” “flaw,” or “faulty interpretation.” In addition, manual searches were performed by screening the citations of review articles and bibliographies of potentially eligible studies. The reference list of included studies, relevant reviews, and authors’ personal files were searched to ensure literature saturation.
STUDY SELECTION AND QUALITY ASSESSMENT:
All eligible articles were downloaded in a portable document format (PDF). The search strategy included a 5-step approach (illustrated in Figure 1). Each paper was assessed regarding potential relevance by screening the titles and abstracts. Subsequently, the full text of articles meeting the eligibility criteria was retrieved and reviewed. Two reviewers (NM and MV) judged the full texts of the potentially eligible reports. If there was a difference in the perceived eligibility of a study, 3 authors (NM, MV, and AB) discussed the report to arrive at a consensus, and the reason for the decision was recorded. We used the PRISMA-P (Preferred Reporting Items for Systematic review and Meta-Analysis Protocols) guidelines to maintain a high-level of quality control throughout the entire study [12].
DATA EXTRACTION AND CLASSIFICATION:
Relevant data from each deficiency frequency report were extracted into a structured spreadsheet. We extracted the quality defect(s) as defined by the author, frequency data (numerator and denominator), sample description, the detection methods used, and citations. Deficiencies were assigned to one of the following deficiency groups; (i) study design; (ii) statistical analysis; (iii) cell lines; or (iv) reporting.
Within each group, identical or essentially similar deficiencies were identified as deficiency concepts (e.g., sample size/power calculation deficiency; mycoplasma contamination of cell lines; or parametric test for non-parametric data). In the groups of study design, statistical analysis, and reporting deficiencies, we used the modified framework of Emerson and Colditz for definition of deficiency concepts [13]. For cell line deficiency concepts, we used the modified framework of both Capes-Davis et al. and Dexler et al. [14,15]. Subsequently, results of collected frequency studies were pooled for meta-analysis based on the deficiency concepts for further meta-analysis.
DATA ANALYSIS:
For deficiency concepts with 4 or more deficiency frequency studies and with a sample size of at least 15 in each, a meta-analysis was performed. Using the Meta-Essentials calculation formulas and software [16], the overall frequency and 95% confidence intervals were calculated for each eligible deficiency concept. The results of this analysis were displayed by multiple forest plots.
To estimate heterogeneity among studies, I2 was used. According to the Cochrane Handbook, heterogeneity is divided into 4 levels: low heterogeneity, 0–25%; moderate heterogeneity, 25–50%; high heterogeneity, 50–75%; and extreme high heterogeneity, 75–100%. Where p<0.05 indicated significant heterogeneity, it could be accepted if the I2 ≤50% [17]. Due the diversity of study sources, we assumed heterogeneity, which was confirmed by the heterogeneity test. The random effects model based on the DerSimonian and Laird approach was used for all studies [18]. Subgroup analysis was performed to explore possible sources of heterogeneity based on the income level of countries, based on the World Bank categorizations [18], which assigns the world’s economies into 4 income groups: high, upper-middle, lower-middle, and low. We combined the upper-middle and lower-middle into one middle category and none of the studies came from the low-income group. We assessed potential regional variation of research quality when sufficient number of deficiencies frequency reports were available for both high-income and middle-income countries.
The publication bias was assessed by funnel plot. Egger regression was used to examine funnel plot asymmetry (p<0.05 indicated significant publication bias). The Begg and Mazumdar rank correlation test was used to examine the funnel plot asymmetry if the deficiency frequency has been published by 10 or more studies [17]. Additionally, the trim and fill method was applied to all forest plots to identify and correct for funnel plot asymmetry arising from publication bias, as well as for estimating the number of missing studies that might exist [19,20]. Accepting recent recommendations, to p-value thresholds were set at 0.005 in this study [21].In addition to the search strategies, all data collected and underlying the findings described in this article are fully available without restriction through Scholarly Commons, the institutional repository for Augusta University (https://augusta.openrepository.com/).
Results
STUDIES OF MULTIPLE SAMPLES AND DEFICIENCIES:
In the pool of eligible studies, 18 reports on deficiency frequency presented results obtained from more than one sample. When a deficiency frequency report was analyzed multiple samples, each sample was given a unique reference number added to the author’s name. For example, the Strasak 2007 study was considered as multiple separate studies and was referenced as Strasak 1 2007, Strasak 2 2007, and so on, for each different sample. Another illustration is the composite publication by Hassan (2015) that reviewed original research studies in multiple groups; therefore, the composite publication was considered a collection of 18 different groups of studies numbered accordingly.
Most deficiency frequency publications analyzed multiple deficiency concepts, not just one using one sample. For example, Lucena [22] estimated the frequency of several study design deficiencies (e.g., eligibility criteria use, power calculation, and randomization) using a sample of 226 dentistry articles.
To illustrate the concept of information aggregation, Figure 2 is an illustrative, partial representation of aggregating studies in the meta-analysis of randomization deficiencies: (a) the left side of the figure shows the level of aggregating information, (b) the middle part shows the pyramid of aggregation from original research studies through deficiency frequency studies and to meta-analysis of deficiency frequency studies, and also the number of studies aggregated and (c) illustrative statements from each level of aggregation.
POOL OF SAMPLES AND DEFICIENCY CONCEPTS:
Of the 68 publications included in this study, several reported the analysis of more than one quality aspect. Ultimately, there were 128 quality aspects analyzed in the collected studies (19 in study design, 63 in cell lines, 18 statistical analysis, and 28 reporting). There were 128 samples and 24 different measured deficiency concepts in the pool of 85 deficiency frequency publications. Based on this information, a total of 24 meta-analyses were performed for quality defects. Deficiency concepts were meta-analyzed in 4 separate groups: study design, cell lines, statistical analysis, and reporting deficiencies.
For the defects in the study design, 3 meta-analyses were conducted based on frequency data provided by 12 research studies that reviewed 1842 original research articles (Figure 3). The deficiency in sample calculation was the most frequent in the study design category, showing an overall frequency of 82.3% [95%: 69.9–94.6%; SE ±6.3%].
Meta-analyses of 6 deficiencies in 64810 cell lines used in life sciences research were analyzed by 42 deficiency frequency studies (Figure 4). The most frequent deficiency was mixed contamination in cell lines, with an overall frequency of 22.4% [95%: 10.4–34.3% SE ±5.3%]. Figure 5 shows the meta-analyses of 3 deficiencies in the statistical analysis of 2419 published research studies provided by 12 deficiency studies. The use of the chi-square test when expected cells frequency was <5 was the most frequent (15.7% [95%: −3.2–34.7%; SE ±7.4%]).
Based on a combined number of 19 studies, 12 meta-analyses were conducted for defects in reporting of 5942 original research results (Figure 6). The most frequent defects were tail numbers not stated, p-values reported without a statistical test, and statistical software not mentioned, showing an overall frequency of 65% [95%: 39.3–90.8%; SE ±10.9%]; 61.5% [95%: 51–72%; SE ±3.8%]; and 54.5% [95%: 34.2–74.9%; SE ±8.6%], respectively.
SUBGROUP ANALYSES:
To investigate the influence of other possible factors on heterogeneity across the studies, subgroup analyses were conducted based on country income level. Table 2 shows the separately estimated I2 variation across studies for both high- and middle-income countries. Our results indicated that there were no significant differences from total variation after separately pooling studies from high- and middle-income countries, except for one deficiency concept. Therefore, based on our results, the country income-based subgroup analysis failed to explain heterogeneity, except for the mean (SD) used for non-normal or ordinal data. Several other subgroup analyses were explored, but none were ultimately feasible due to the insufficient number of error frequency reports (e.g., year of publications, cell line type non-cancer/cancer, human/animal/mixed). In other words, there was a lack of evidence for quality improvement over time.
Analysis of publication bias showed funnel plot symmetry and corresponding lack of statistically significant bias for the majority of studied deficiency concepts. There were some exceptions, suggesting publication bias in the literature: cell line bacterial contamination other than mycoplasma (p=0.002); mixed contamination of cell lines (p=0.002); chi-square test used when expected cells frequency <5 (p=0.007); and p-value significance level not defined (p=0.007). (Figure 7).
Discussion
While research is inherently innovative and variable, many methodologies became routinely used; therefore, associated deficiency rates are increasingly recognized. Due to the growing number of deficiency frequency studies, integration of results is becoming possible and necessary. This series of meta-analyses is the first comprehensive study to provide numeric frequency estimates for 21 different deficiencies in life sciences research.
The complexity of the life sciences research process makes it prone to deficiencies. Some research studies use pioneering or unique methodologies, but many studies use standard methods (e.g., knockout mouse, standard cell lines). Results of this study indicate that the frequency of deficiencies in life sciences research can be reliably measured.
Interestingly, the studies on possible reasons for non-reproducibility have been largely based on expert opinion and are themselves non-reproducible. This study represents the first comprehensive collection of research deficiency detection studies that solely relies on deficiency definitions successfully reproduced in several studies.
We found that deficiency rates vary between 1.3% and 82.3%, depending on the particular type of deficiency in life sciences research. Our comprehensive meta-analysis indicates that the following deficiencies in life sciences research are particularly frequent (i.e., meta-frequency exceeding 20%): sample size/power calculation deficiency, tails number not stated, p-values reported without statistical test, statistical software not mentioned, eligibility criteria incomplete, failure to report the exact p-value, p-value significance level not defined, randomization deficiency, statistical test used for a dataset not specified, mixed contamination of cell lines, and no description of the study population.
When many researchers use at least partly identical methodologies, certain deficiencies are becoming recognizable and their frequencies can be estimated. This does not mean that the particular methodology is flawed, only that it is vulnerable to certain deficiencies. For example, the use of cancer cell lines is an excellent laboratory methodology, but it is occasionally vulnerable to misidentification or contamination. Researchers need to be aware of such sources of deficiencies and prepared to prevent and detect them.
Scientific quality control has long been reliant on peer review. However, such control is too late when the research itself is already done. For many defects, it would be more advantageous to consider them while the research is still progressing. This meta-analysis provides actionable and measurable defect identification, unlike the majority of articles on quality control in research. When scientists get these numbers, they should know which errors are more frequent and what needs to be considered at a particular phase of their study.
This study focused on 4 key research aspects relevant to the reproducibility of results from the initial to the late phases of basic biomedical research (study design, cell lines, statistical analysis, and reporting). This information should be valuable for researchers and also research administrators in recognizing the most frequent errors and to prevent them most effectively. As new aggregate research deficiency studies will be emerging, they can be added to expand the scope and applicability to quality improvement in research laboratories and institutions.
The deficiencies highlighted by this meta-analysis were the most frequent within their own category (e.g., cell line contamination). It should also be recognized that the deficiencies reported were not necessarily the most important sources of irreproducibility either during the review period or for the present time. There might be other errors that have not been systematically measured yet but that can be included in the future when pertinent frequency measurements arise. Our study should encourage further and wider-ranging studies on the frequency of deficiencies of biomedical research.
This study did not find evidence that variations in the frequency of research reproducibility deficiencies are explained by differences between high-income and middle-income countries. Apparently, the income environment does not influence the quality of research, although it may influence the choice of research focus and access to resources. There are many distinguished scientists from the developing world who are making important contributions to the scientific community worldwide.
It is well recognized that the number of scientific publications is rapidly growing worldwide. The rising trends of research publications can be partly attributed to the increase of international scientific collaboration. Researchers, funders, and journal editors communicate science the same way all over the world. The method of science has to meet the same quality standards everywhere and is not linked to the region.
The potential for quality improvement over time was considered, but we found no evidence of such trends. It is possible that the timeframe of available data-driven quality studies was not sufficient to detect changes/improvement over time.
The lack of evidence for research quality improvement over time is not surprising, for several reasons. The potential for quality improvement over time was not the scope of this analysis, as the included studies have different methodologies and sample types, making comparisons difficult. According to the principles of management science, general improvement in quality comes from systematic and regular measurements of deficiencies and organized efforts to manage quality (e.g., car manufacturing industry, health care quality improvement in many countries). With rare exceptions, such systematic institutional quality management initiatives are uncommon in the biomedical research enterprise.
A limitation of the present study is the reliance on already-published numeric analyses of research deficiencies. There are many more suspected and actual life sciences research deficiencies that have not yet been analyzed by a sufficient number of studies to be included in this meta-analysis (e.g., dysfunctional reagents). Further, subgroup analysis by sample type was not possible due to insufficient sample size. It is also obvious that defects in research are probably under reported. Moreover, in the cell line group, different studies used different techniques for identifying the various defects in cell lines. Our study selection was restricted to articles published in English. It is possible that studies published in other languages or unpublished studies could shift the overall conclusion.
The 4 deficiency categories were selected based on reviewing the literature, talking to scientists in the field, and the repeated NIH calls to enhancing reproducibility and integrity of research [23]. Deficiency in animal studies was one of these categories. In collecting studies for these meta-analyses, several highly publicized articles on research quality issues did not provide frequency estimates and thus were not eligible for inclusion. Due to the diversity of issues and defects, animal modeling studies was not the target of our study. While this series of meta-analyses was intentionally comprehensive, it should not be considered all-inclusive, as the list of recognized research quality deficiencies is continuously evolving.
Management science often stresses that narratives without data are rarely effective in improving quality. This meta-analysis shows the theoretical and practical significance of measuring quality in basic biomedical research. With more emphasis on continuous quality improvement, the number of deficiency frequency studies is likely to substantially grow.
Conclusions
Research quality improvement should be a continuous and comprehensive process, from the design and conduct of research to the publication of results. With periodic analyses, corrective actions should be recommended and implemented to reduce the chances of deficiencies. Life sciences research deficiencies can be one of the following types.
Continuous quality improvement is a major challenge that needs to be fully recognized by research institutes and universities. A collaborative culture at the institutional level is needed to eliminate deficiencies in life sciences research. Researchers and research institutions need to appreciate the value of measurement of deficiencies and work together to implement the needed changes. Improvement efforts should be built on these comprehensive measures, which should reduce deficiencies, increase research productivity, and multiply meritorious scientific discoveries.
Figures
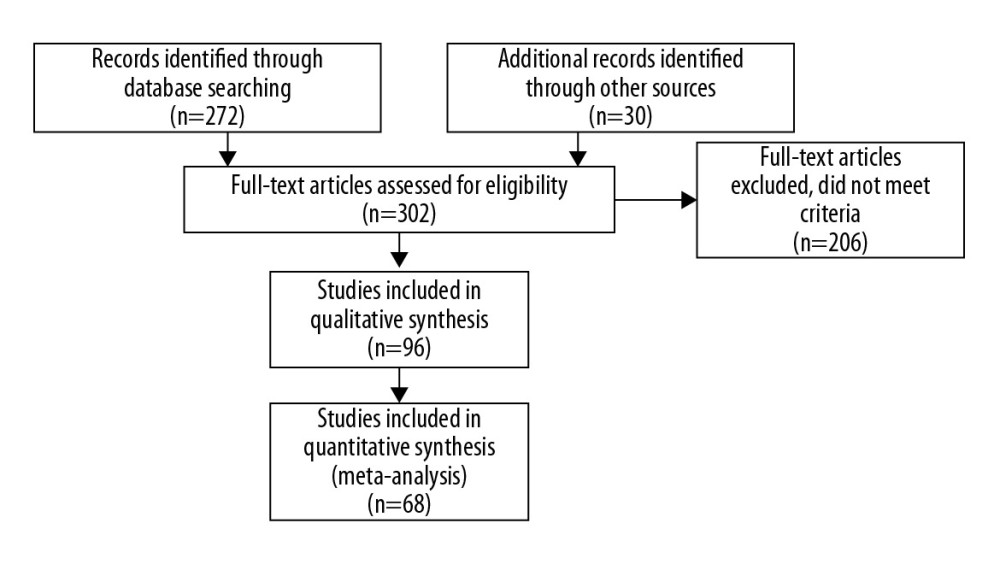 Figure 1. Search Strategy of the Meta-Analysis. (PRISMA, 2009).
Figure 1. Search Strategy of the Meta-Analysis. (PRISMA, 2009). 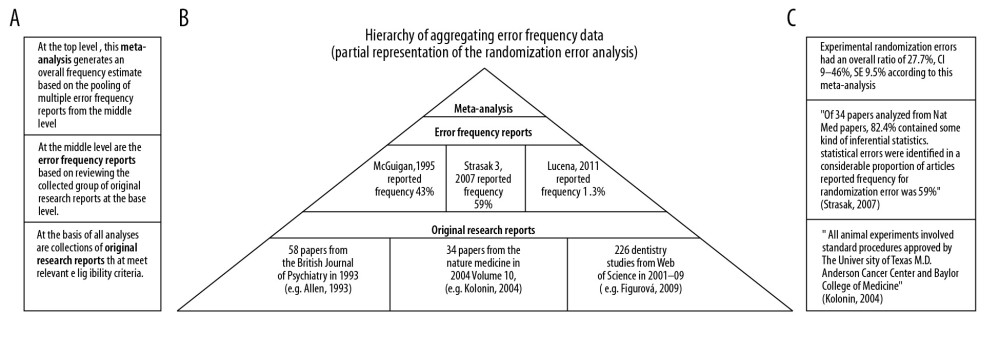 Figure 2. Simplified illustration of the aggregation of information. (A) Description of study levels; (B) pyramid of aggregating information about research deficiencies; and (C) illustrative study statements at each level.
Figure 2. Simplified illustration of the aggregation of information. (A) Description of study levels; (B) pyramid of aggregating information about research deficiencies; and (C) illustrative study statements at each level. 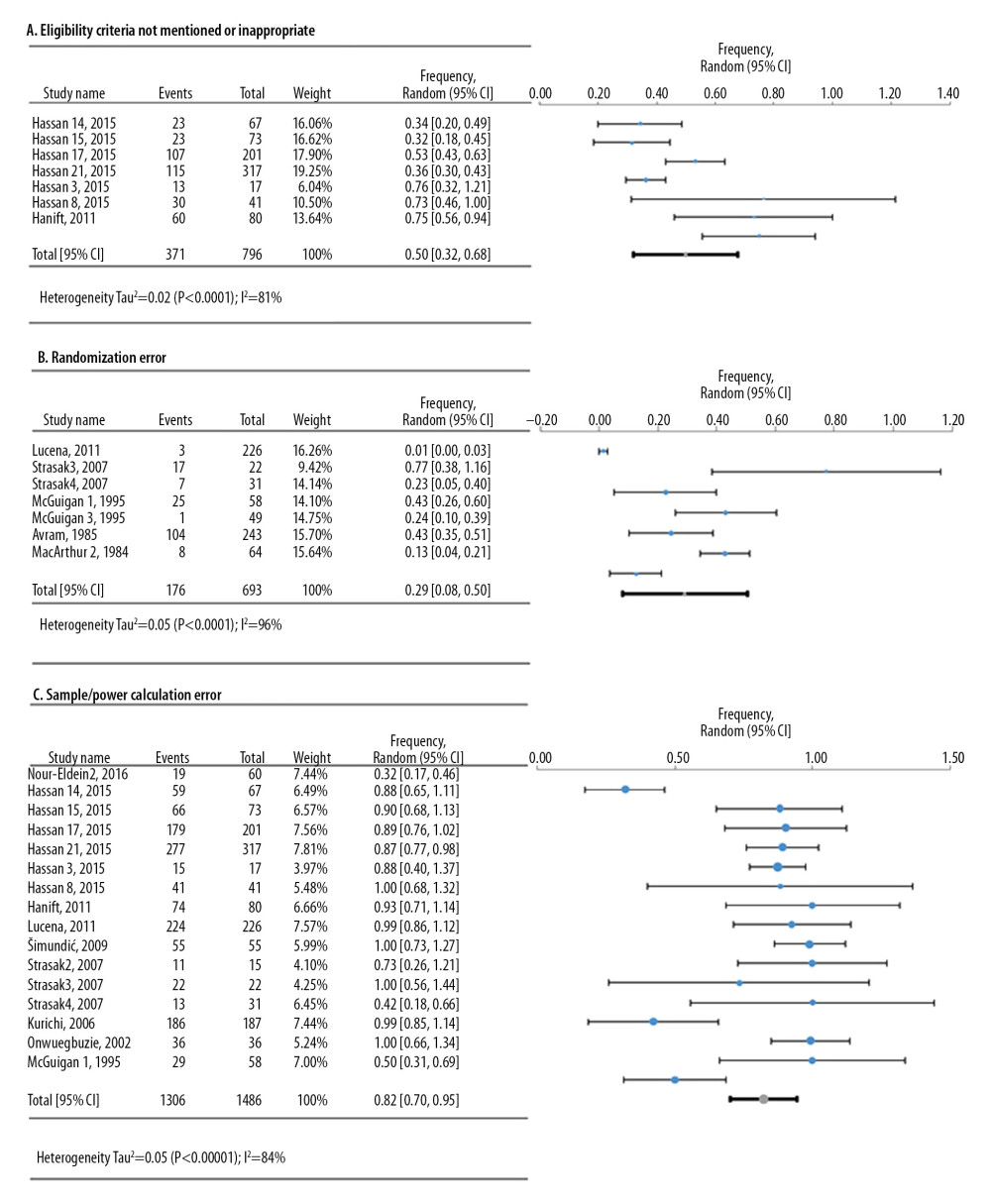 Figure 3. Frequency estimates of 3 (A–C) study design deficiencies in original research articles.
Figure 3. Frequency estimates of 3 (A–C) study design deficiencies in original research articles. 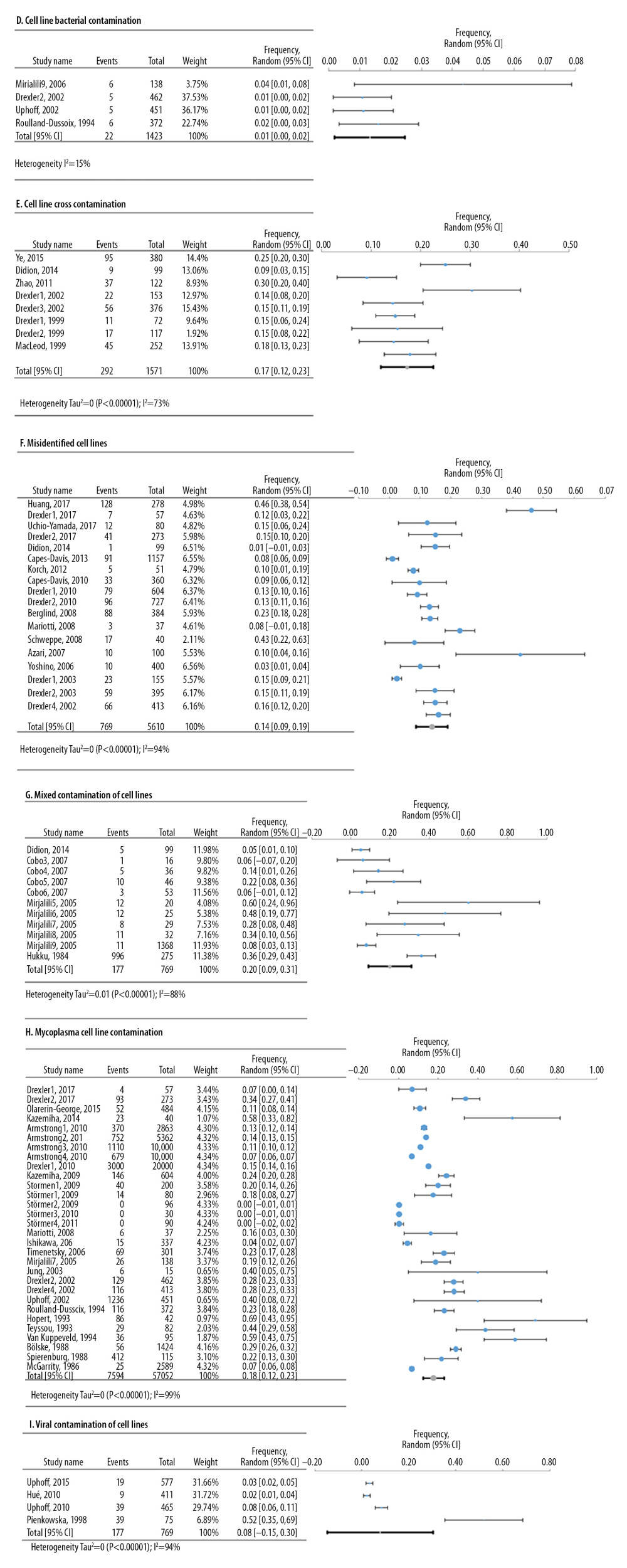 Figure 4. Frequency estimates of 6 (D–I) cell line defects.
Figure 4. Frequency estimates of 6 (D–I) cell line defects. 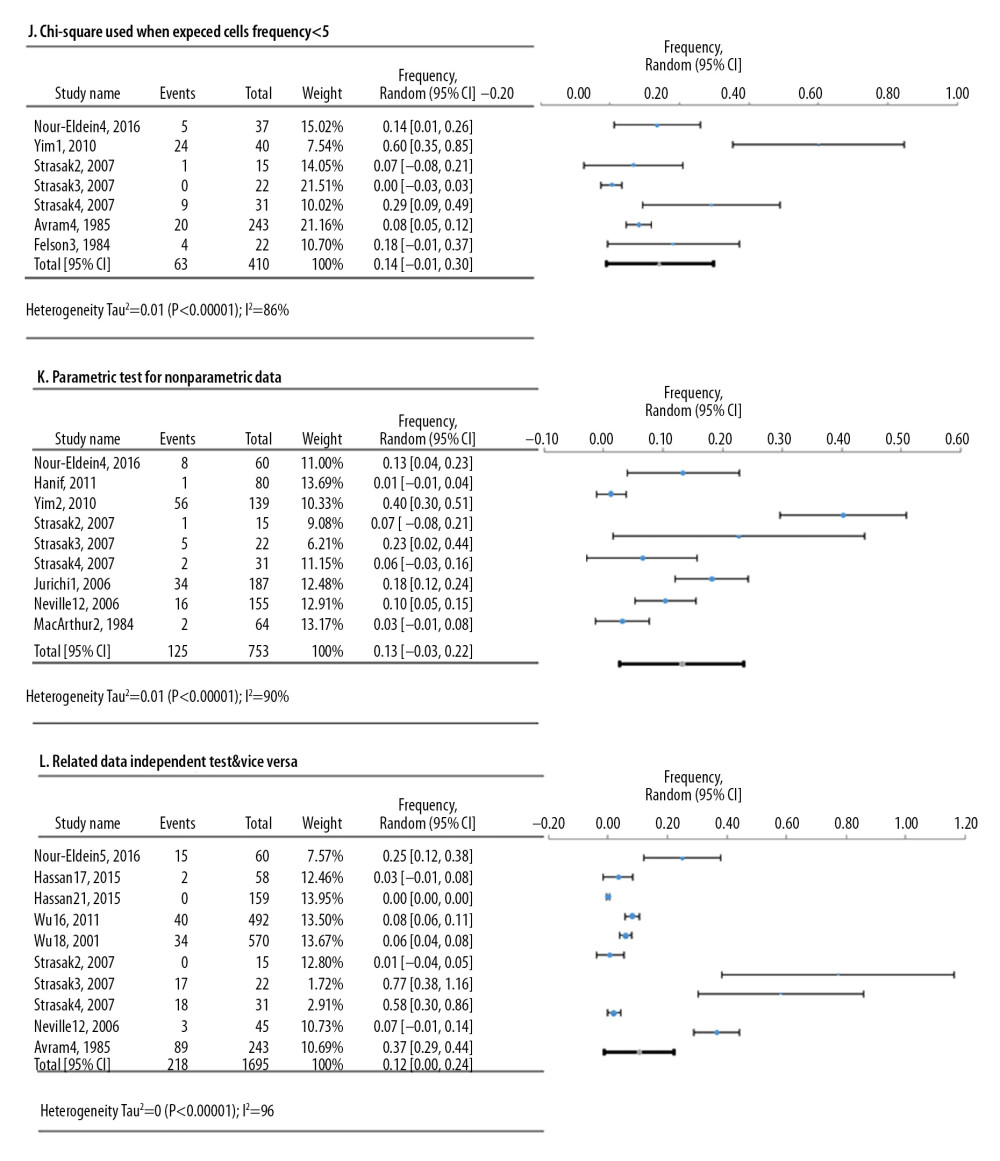 Figure 5. Frequency estimates of 3 (J–L) statistical analysis deficiencies in original research articles.
Figure 5. Frequency estimates of 3 (J–L) statistical analysis deficiencies in original research articles. 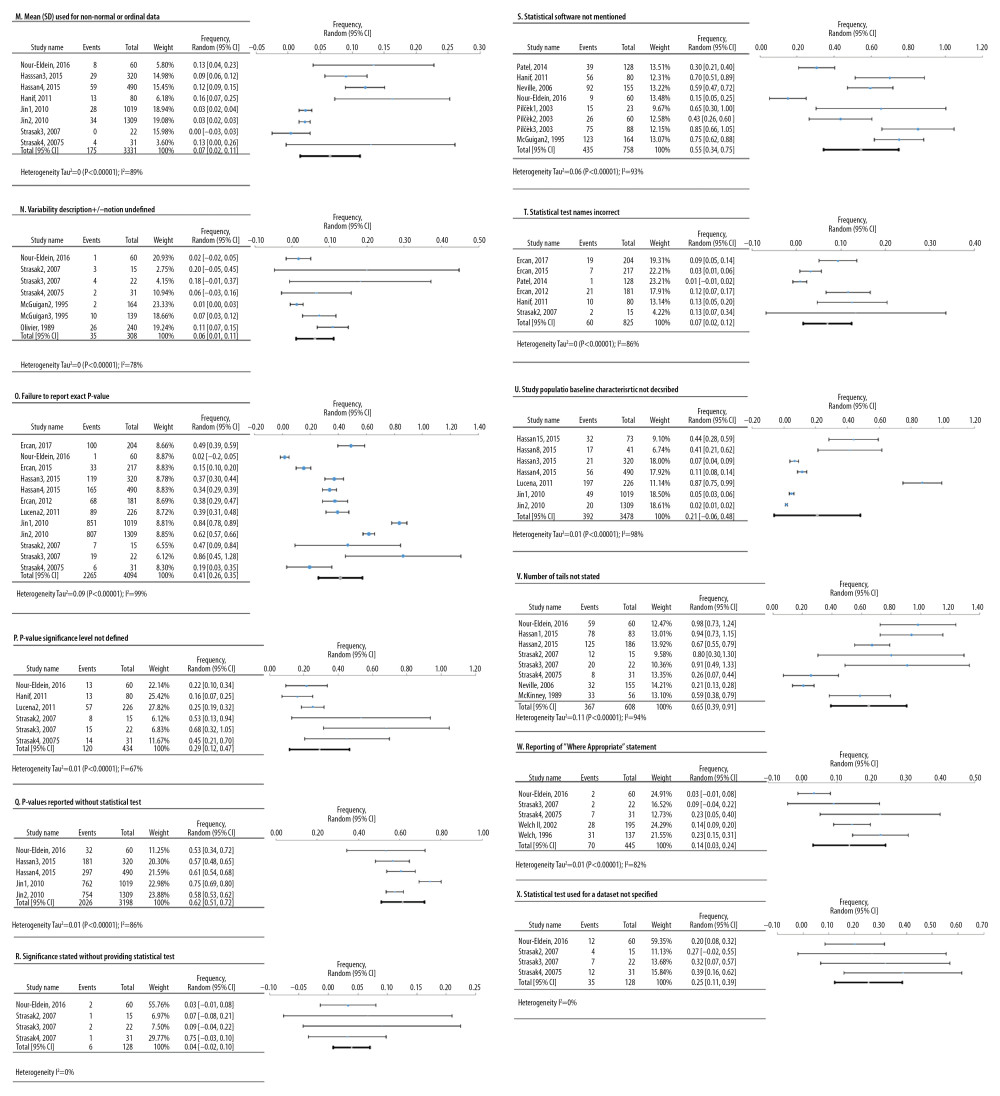 Figure 6. Frequency estimates of 12 (M–X) reporting deficiencies in original research articles.
Figure 6. Frequency estimates of 12 (M–X) reporting deficiencies in original research articles. 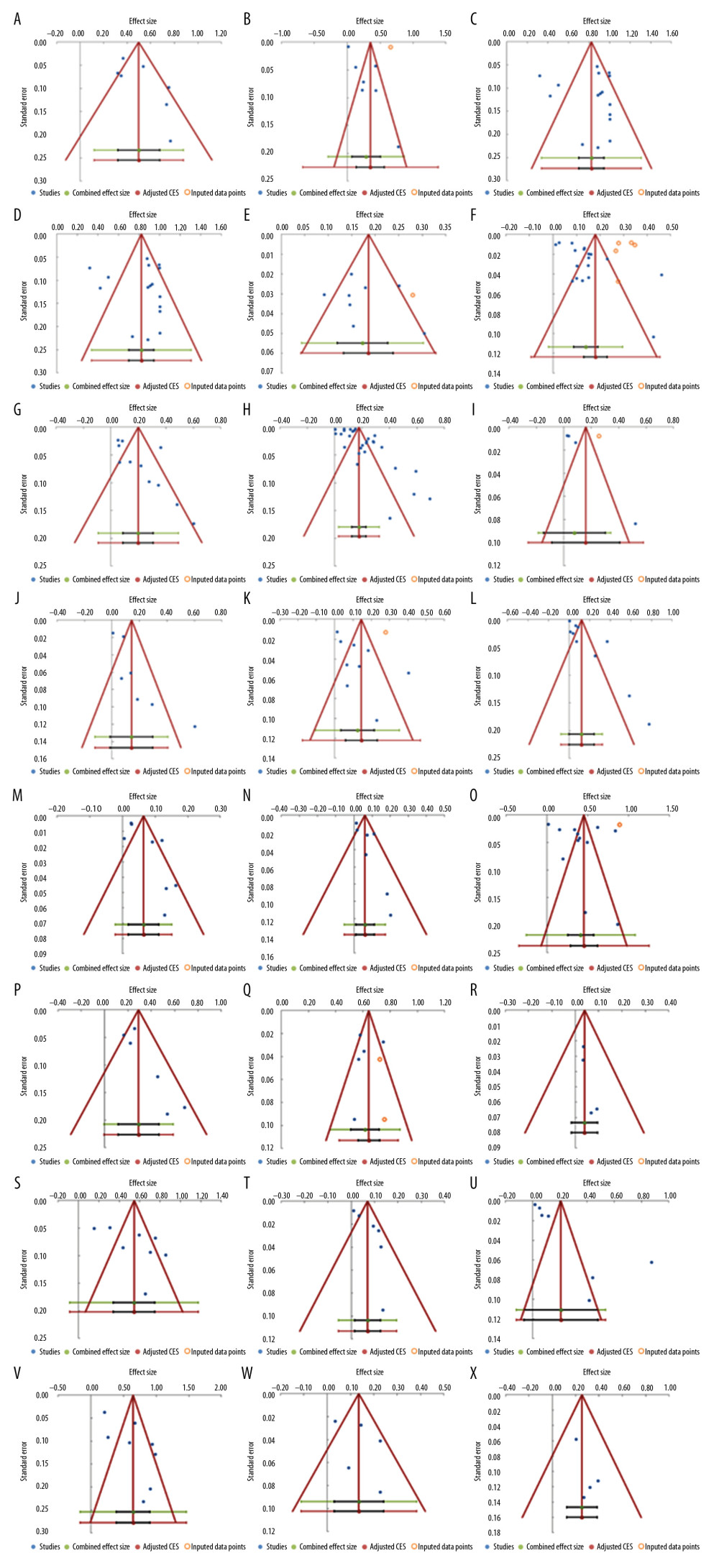 Figure 7. Funnel plots. (A) Eligibility criteria not mentioned or inappropriate (B) Randomization deficiency (C) Sample/power calculation deficiency (D) Cell line bacterial contamination other than mycoplasma (E). Cell line cross-contamination (F). Misidentified cell lines (G). Mixed contamination of cell lines (H). Mycoplasma cell line contamination (I). Viral contamination of cell lines (J). Chi-square test used when expected cells frequency are <5 (K). Parametric test for non-parametric data and vice versa (L). Related data independent test and vice versa (M). Mean(SD) used for non-normal or ordinal data (N). variability description +/− notation undefined (O). Failure to report exact p-value (P). p-value significance level not defined (Q). p-value reported without statistical test (R). Significance stated without providing statistical test (S). Statistical software not mentioned (T). Statistical test name incorrect (U). Study population baseline characteristics not described (V). Number of tails not stated (W). Reporting of “Where appropriate statement” (X). Statistical test used for dataset not specified.
Figure 7. Funnel plots. (A) Eligibility criteria not mentioned or inappropriate (B) Randomization deficiency (C) Sample/power calculation deficiency (D) Cell line bacterial contamination other than mycoplasma (E). Cell line cross-contamination (F). Misidentified cell lines (G). Mixed contamination of cell lines (H). Mycoplasma cell line contamination (I). Viral contamination of cell lines (J). Chi-square test used when expected cells frequency are <5 (K). Parametric test for non-parametric data and vice versa (L). Related data independent test and vice versa (M). Mean(SD) used for non-normal or ordinal data (N). variability description +/− notation undefined (O). Failure to report exact p-value (P). p-value significance level not defined (Q). p-value reported without statistical test (R). Significance stated without providing statistical test (S). Statistical software not mentioned (T). Statistical test name incorrect (U). Study population baseline characteristics not described (V). Number of tails not stated (W). Reporting of “Where appropriate statement” (X). Statistical test used for dataset not specified. References
1. Freedman LP, Cockburn IM, Simcoe TS, The economics of reproducibility in preclinical research: PLoS Biol, 2015; 13(6); e1002165
2. Begley CG, Ellis LM, Drug development: Raise standards for preclinical cancer research: Nature, 2012; 483(7391); 531-33
3. Bespalov A, Barnett A, Begley C, Industry is more alarmed about reproducibility than academia: Nature, 2018; 563(7733); 626
4. Ioannidis JP, Why most published research findings are false: PLoS Med, 2005; 2(8); e124
5. Karp NA, Reproducible preclinical research – Is embracing variability the answer?: PLoS Biol, 2018; 16(3); e2005413
6. Naik G, Mistakes in scientific studies surge: Wall St J, 2011; 10
7. Prinz F, Schlange T, Asadullah K, Believe it or not: How much can we rely on published data on potential drug targets?: Nat Rev Drug Discov, 2011; 10(9); 712
8. Casadevall A, Steen RG, Fang FC, Sources of error in the retracted scientific literature: FASEB J, 2014; 28(9); 3847-55
9. Foster JG, Rzhetsky A, Evans JA, Tradition and innovation in scientists’ research strategies: American Sociological Review, 2015; 80(5); 875-908
10. Cottler LB, Zunt J, Weiss B, Building global capacity for brain and nervous system disorders research: Nature, 2015; 527(7578); S207-13
11. Collins FS, Tabak LA, Policy: NIH plans to enhance reproducibility: Nature, 2014; 505(7485); 612-13
12. Shamseer L, Moher D, Clarke M, Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: Elaboration and explanation: BMJ, 2015; 349; g7647
13. Emerson JD, Colditz GA, Use of statistical analysis in the New England Journal of Medicine: N Engl J Med, 1983; 309(12); 709-13
14. Capes-Davis A, Theodosopoulos G, Atkin I, Check your cultures! A list of cross-contaminated or misidentified cell lines: Int J Cancer, 2010; 127(1); 1-8
15. Drexler HG, Uphoff CC, Dirks WG, MacLeod RA, Mix-ups and mycoplasma: The enemies within: Leuk Res, 2002; 26(4); 329-33
16. van Rhee H, Suurmond R, Hak T: User manual for Meta-Essentials: Workbooks for meta-analyses (Version 1.0), 2015
17. Higgins JP, Green S: Cochrane handbook for systematic reviews of interventions. Version, 2005
18. Bank W: World Bank Country and Lending Groups, 2018
19. Taylor S, Weedie TR: A simple funnel plot based method of testing and adjusting for publication bias in meta-analyses, 1998, Fort Collins, CO, Colorado State University
20. Duval S, Tweedie R, A nonparametric “trim and fill” method of accounting for publication bias in meta-analysis: Journal of the American Statistical Association, 2000; 95(449); 89-98
21. Ioannidis JP, The proposal to lower P value thresholds to. 005: JAMA, 2018; 319(14); 1429-30
22. Lucena C, Lopez JM, Abalos C, Statistical errors in microleakage studies in operative dentistry. A survey of the literature 2001–2009: Eur J Oral Sci, 2011; 119(6); 504-10
23. NIH: Enhancing Reproducibility in NIH Applications: Resource Chart [January 25, 2019] Available from: https://grants.nih.gov/grants/Rigor-and-Reproducibility-Chart-508.pdf
24. Benjamin DJ, Berger JO, Johannesson M, Redefine statistical significance: Nat Hum Behav, 2018; 2(1); 6-10
25. Armstrong SE, Mariano JA, Lundin DJ, The scope of mycoplasma contamination within the biopharmaceutical industry: Biologicals, 2010; 38(2); 211-13
26. Mariotti E, Mirabelli P, Di Noto R, Rapid detection of mycoplasma in continuous cell lines using a selective biochemical test: Leuk Res, 2008; 32(2); 323-26
27. Avram MJ, Shanks CA, Dykes MH, Statistical methods in anesthesia articles: An evaluation of two American journals during two six-month periods: Anesth Analg, 1985; 64(6); 607-11
28. McGarrity GJ, Kotani H, Carson D, Comparative studies to determine the efficiency of 6 methylpurine deoxyriboside to detect cell culture mycoplasmas: In vitro Cell Dev Biol, 1986; 22(6); 301-4
29. Azari S, Ahmadi N, Tehrani MJ, Shokri F: Biologicals, 2007; 35(3); 195-202
30. McGuigan SM, The use of statistics in the British Journal of Psychiatry: Br J Psychiatry, 1995; 167(5); 683-88
31. Berglind H, Pawitan Y, Kato S, Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination: Cancer Biol Ther, 2008; 7(5); 699-708
32. McKinney WP, Young MJ, Hartz A, Lee M, The inexact use of Fisher’s exact test in six major medical journals: JAMA, 1989; 261(23); 3430-33
33. Bolske G, Survey of Mycoplasma infections in cell cultures and a comparison of detection methods: Zentralbl Bakteriol Mikrobiol Hyg A, 1988; 269(3); 331-40
34. Mirjalili A, Parmoor E, Bidhendi SM, Sarkari B, Microbial contamination of cell cultures: A 2 years study: Biologicals, 2005; 33(2); 81-85
35. Neville JA, Lang W, Fleischer AB, Errors in the Archives of Dermatology and the Journal of the American Academy of Dermatology from January through December 2003: Arch Dermatol, 2006; 142(6); 737-40
36. Capes-Davis A, Reid YA, Kline MC, Match criteria for human cell line authentication: where do we draw the line?: Int J Cancer, 2013; 132(11); 2510-19
37. Nour-Eldein H, Statistical methods and errors in family medicine articles between 2010 and 2014 – Suez Canal University, Egypt: A cross-sectional study: J Family Med Prim Care, 2016; 5(1); 24-33
38. Cobo F, Cortes JL, Cabrera C, Microbiological contamination in stem cell cultures: Cell Biol Int, 2007; 31(9); 991-95
39. Olarerin-George AO, Hogenesch JB, Assessing the prevalence of mycoplasma contamination in cell culture via a survey of NCBI’s RNA-seq archive: Nucleic Acids Res, 2015; 43(5); 2535-42
40. Didion JP, Buus RJ, Naghashfar Z, SNP array profiling of mouse cell lines identifies their strains of origin and reveals cross-contamination and widespread aneuploidy: BMC Genomics, 2014; 15; 847
41. Oliver D, Hall J, Usage of statistics in the surgical literature and the ‘orphan P’phenomenon: Aust NZ J Surg, 1989; 59(6); 449-51
42. Drexler H, Dirks W, MacLeod R, False human hematopoietic cell lines: Cross-contaminations and misinterpretations: Leukemia, 1999; 13(10); 1601
43. Onwuegbuzie AJ, Common methodological, analytical, and interpretational errorsin published educational studies: An analysis of the 1998 volume of the British Journal of Educational Psychology: Educational Research Quarterly, 2002; 26(1); 11
44. Patel S, Naik V, Patel P, Use of statistical methods and complexity of data analysis in recent research publications in basic medical sciences: Community Med, 2014; 5(2); 253-56
45. Drexler H, Dirks W, Matsuo Y, MacLeod R, False leukemia–lymphoma cell lines: An update on over 500 cell lines: Leukemia, 2003; 17(2); 416-26
46. Pienkowska M, Seth A, Detection of squirrel monkey retroviral sequences in interferon samples: J Hepatol, 1998; 28(3); 396-403
47. Drexler HG: Guide to Leukemia-Lymphoma Cell Lines Braunschweig, 2010; 883-92
48. Pilčèk T, Statistics in three biomedical journals: Physiol Res, 2003; 52; 39-43
49. Drexler HG, Dirks WG, MacLeod RA, Uphoff CC, False and mycoplasma-contaminated leukemia-lymphoma cell lines: time for a reappraisal: Int J Cancer, 2017; 140(5); 1209-14
50. Roulland-Dussoix D, Henry A, Lemercier B, Detection of mycoplasmas in cell cultures by PCR: A one year study: J Microbiol Methods, 1994; 19(2); 127-34
51. Ercan I, Ocakoğlu G, Siğirli D, Özkaya G, Assessment of submitted manuscripts in medical sciences according to statistical errors: Turkiye Klinikleri Journal of Medical Sciences, 2012; 3 2(5); 1381-87
52. Schweppe RE, Klopper JP, Korch C, Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification: J Clin Endocrinol Metab, 2008; 93(11); 4331-41
53. Ercan I, Karadeniz PG, Cangur S, Examining of published articles with respect to statistical errors in medical sciences: International Journal of Hematology and Oncology, 2015; 27(4); 130-38
54. Šimundić A-M, Nikolac N, Statistical errors in manuscripts submitted to Biochemia Medica journal: Biochemia Medica, 2009; 19(3); 294-300
55. Ercan I, Kaya MO, Uzabaci E, Examination of published articles with respect to statistical errors in veterinary sciences: Acta Veterinaria, 2017; 67(1); 33-42
56. Spierenburg GT, Polak-Vogelzang AA, Bast BJ, Indicator cell lines for the detection of hidden mycoplasma contamination, using an adenosine phosphorylase screening test: J Immunol Methods, 1988; 114(1–2); 115-19
57. Felson DT, Adrienne Cupples L, Meenan RF, Misuse of statistical methods in arthritis and rheumatism: Arthritis Rheum, 1984; 27(9); 1018-22
58. Stormer M, Vollmer T, Henrich B, Broad-range real-time PCR assay for the rapid identification of cell-line contaminants and clinically important mollicute species: Int J Med Microbiol, 2009; 299(4); 291-300
59. Hanif A, Ajmal T, Statistical errors in medical journals (A critical appraisal): Annals of King Edward Medical University, 2011; 17(2); 178
60. Strasak AM, Zaman Q, Marinell G, The use of statistics in medical research: A comparison of The New England Journal of Medicine and Nature Medicine: The American Statistician, 2007; 61(1); 47-55
61. Hassan S, Yellur R, Subramani P, Research design and statistical methods in Indian medical journals: a retrospective survey: PLoS One, 2015; 10(4); e0121268
62. Strasak AM, Zaman Q, Marinell G, The use of statistics in medical research: A comparison of Wiener Klinische Wochenschrift andWiener Medizinische Wochenschrift: Austrian Journal of Statistics, 2007; 36(2); 141-52
63. Hopert A, Uphoff CC, Wirth M, Specifity and sensitivity of polymerase chain reaction (PCR) in comparison with other methods for the detection of mycoplasma contamination in cell lines: J Immunol Methods, 1993; 164(1); 91-100
64. Teyssou R, Poutiers F, Saillard C, Detection of mollicute contamination in cell cultures by 16S rDNA amplification: Mol Cell Probes, 1993; 7(3); 209-16
65. Huang Y, Liu Y, Zheng C, Shen C, Investigation of cross-contamination and misidentification of 278 widely used tumor cell lines: PLoS One, 2017; 12(1); e0170384
66. Timenetsky J, Santos L, Buzinhani M, Mettifogo E, Detection of multiple mycoplasma infection in cell cultures by PCR: Braz J Med Biol Res, 2006; 39(7); 907-14
67. Hué S, Gray ER, Gall A, Disease-associated XMRV sequences are consistent with laboratory contamination: Retrovirology, 2010; 7(1); 111
68. Uchio-Yamada K, Kasai F, Ozawa M, Kohara A, Incorrect strain information for mouse cell lines: Sequential influence of misidentification on sublines: In Vitro Cell Dev Biol Anim, 2017; 53(3); 225-30
69. Hukku B, Halton DM, Mally M, Peterson WD, Cell characterization by use of multiple genetic markers: Adv Exp Med Biol, 1984; 172; 13-31
70. Uphoff C, Drexler H, Detection of mycoplasma in leukemia–lymphoma cell lines using polymerase chain reaction: Leukemia, 2002; 16(2); 289-93
71. Ishikawa Y, Kozakai T, Morita H, Rapid detection of mycoplasma contamination in cell cultures using SYBR Green-based real-time polymerase chain reaction: In Vitro Cell Dev Biol Anim, 2006; 42(3–4); 63-69
72. Uphoff CC, Denkmann SA, Steube KG, Drexler HG, Detection of EBV, HBV, HCV, HIV-1, HTLV-I and-II, and SMRV in human and other primate cell lines: J Biomed Biotechnol, 2010; 2010 904767
73. Jin Z, Yu D, Zhang L, A retrospective survey of research design and statistical analyses in selected Chinese medical journals in 1998 and 2008: PLoS One, 2010; 5(5); e10822
74. Uphoff CC, Lange S, Denkmann SA, Prevalence and characterization of murine leukemia virus contamination in human cell lines: PLoS One, 2015; 10(4); e0125622
75. Jung H, Wang SY, Yang IW, Detection and treatment of mycoplasma contamination in cultured cells: Chang Gung Med J, 2003; 26(4); 250-58
76. Van Kuppeveld F, Johansson K, Galama J, Detection of mycoplasma contamination in cell cultures by a mycoplasma group-specific PCR: Appl Environ Microbiol, 1994; 60(1); 149-52
77. Kazemiha VM, Shokrgozar MA, Arabestani MR, PCR-based detection and eradication of mycoplasmal infections from various mammalian cell lines: A local experience: Cytotechnology, 2009; 61(3); 117-24
78. Welch GE, Gabbe SG, Statistics usage in the American Journal of Obstetrics and Gynecology: has anything changed?: Am J Obstet Gynecol, 2002; 186(3); 584-86
79. Kazemiha VM, Amanzadeh A, Memarnejadian A, Sensitivity of biochemical test in comparison with other methods for the detection of mycoplasma contamination in human and animal cell lines stored in the National Cell Bank of Iran: Cytotechnology, 2014; 66(5); 861-73
80. Welch GE, Gabbe SG, Review of statistics usage in the American Journal of Obstetrics and Gynecology: Am J Obstet Gynecol, 1996; 175(5); 1138-41
81. Korch C, Spillman MA, Jackson TA, DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination: Gynecol Oncol, 2012; 127(1); 241-48
82. Wu S, Jin Z, Wei X, Misuse of statistical methods in 10 leading Chinese medical journals in 1998 and 2008: ScientificWorldJournal, 2011; 11; 2106-14
83. Kurichi JE, Sonnad SS, Statistical methods in the surgical literature: J Am Coll Surg, 2006; 202(3); 476-84
84. Ye F, Chen C, Qin J, Genetic profiling reveals an alarming rate of cross-contamination among human cell lines used in China: FASEB J, 2015; 29(10); 4268-72
85. Yim KH, Nahm FS, Han KA, Park SY, Analysis of statistical methods and errors in the articles published in the korean journal of pain: Korean J Pain, 2010; 23(1); 35-41
86. MacArthur RD, Jackson GG, An evaluation of the use of statistical methodology in the Journal of Infectious Diseases: J Infect Dis, 1984; 149(3); 349-54
87. Yoshino K, Iimura E, Saijo K, Essential role for gene profiling analysis in the authentication of human cell lines: Hum Cell, 2006; 19(1); 43-48
88. MacLeod RA, Dirks WG, Matsuo Y, Widespread intraspecies cross-contamination of human tumor cell lines arising at source: Int J Cancer, 1999; 83(4); 555-63
89. Zhao M, Sano D, Pickering CR, Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites: Clin Cancer Res, 2011; 17(23); 7248-64
Figures
 Figure 1. Search Strategy of the Meta-Analysis. (PRISMA, 2009).
Figure 1. Search Strategy of the Meta-Analysis. (PRISMA, 2009). Figure 2. Simplified illustration of the aggregation of information. (A) Description of study levels; (B) pyramid of aggregating information about research deficiencies; and (C) illustrative study statements at each level.
Figure 2. Simplified illustration of the aggregation of information. (A) Description of study levels; (B) pyramid of aggregating information about research deficiencies; and (C) illustrative study statements at each level. Figure 3. Frequency estimates of 3 (A–C) study design deficiencies in original research articles.
Figure 3. Frequency estimates of 3 (A–C) study design deficiencies in original research articles. Figure 4. Frequency estimates of 6 (D–I) cell line defects.
Figure 4. Frequency estimates of 6 (D–I) cell line defects. Figure 5. Frequency estimates of 3 (J–L) statistical analysis deficiencies in original research articles.
Figure 5. Frequency estimates of 3 (J–L) statistical analysis deficiencies in original research articles. Figure 6. Frequency estimates of 12 (M–X) reporting deficiencies in original research articles.
Figure 6. Frequency estimates of 12 (M–X) reporting deficiencies in original research articles. Figure 7. Funnel plots. (A) Eligibility criteria not mentioned or inappropriate (B) Randomization deficiency (C) Sample/power calculation deficiency (D) Cell line bacterial contamination other than mycoplasma (E). Cell line cross-contamination (F). Misidentified cell lines (G). Mixed contamination of cell lines (H). Mycoplasma cell line contamination (I). Viral contamination of cell lines (J). Chi-square test used when expected cells frequency are <5 (K). Parametric test for non-parametric data and vice versa (L). Related data independent test and vice versa (M). Mean(SD) used for non-normal or ordinal data (N). variability description +/− notation undefined (O). Failure to report exact p-value (P). p-value significance level not defined (Q). p-value reported without statistical test (R). Significance stated without providing statistical test (S). Statistical software not mentioned (T). Statistical test name incorrect (U). Study population baseline characteristics not described (V). Number of tails not stated (W). Reporting of “Where appropriate statement” (X). Statistical test used for dataset not specified.
Figure 7. Funnel plots. (A) Eligibility criteria not mentioned or inappropriate (B) Randomization deficiency (C) Sample/power calculation deficiency (D) Cell line bacterial contamination other than mycoplasma (E). Cell line cross-contamination (F). Misidentified cell lines (G). Mixed contamination of cell lines (H). Mycoplasma cell line contamination (I). Viral contamination of cell lines (J). Chi-square test used when expected cells frequency are <5 (K). Parametric test for non-parametric data and vice versa (L). Related data independent test and vice versa (M). Mean(SD) used for non-normal or ordinal data (N). variability description +/− notation undefined (O). Failure to report exact p-value (P). p-value significance level not defined (Q). p-value reported without statistical test (R). Significance stated without providing statistical test (S). Statistical software not mentioned (T). Statistical test name incorrect (U). Study population baseline characteristics not described (V). Number of tails not stated (W). Reporting of “Where appropriate statement” (X). Statistical test used for dataset not specified. Tables
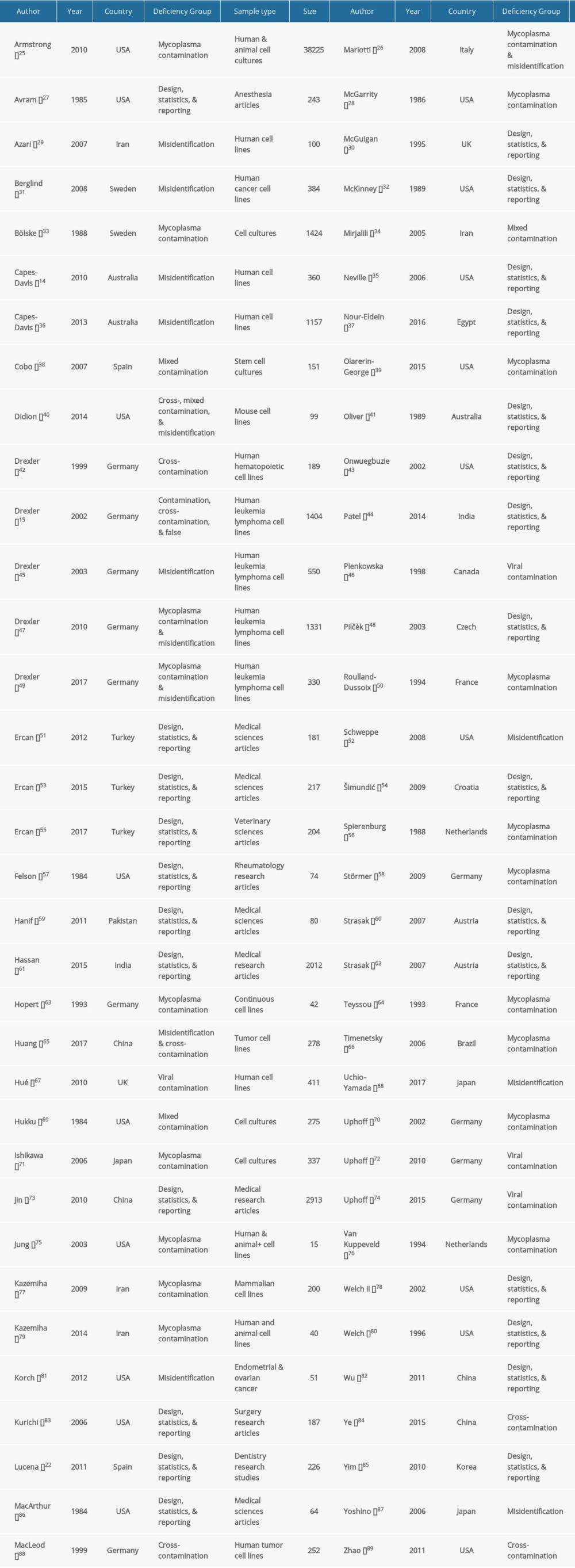 Table 1. Baseline characteristics of the included studies.
Table 1. Baseline characteristics of the included studies.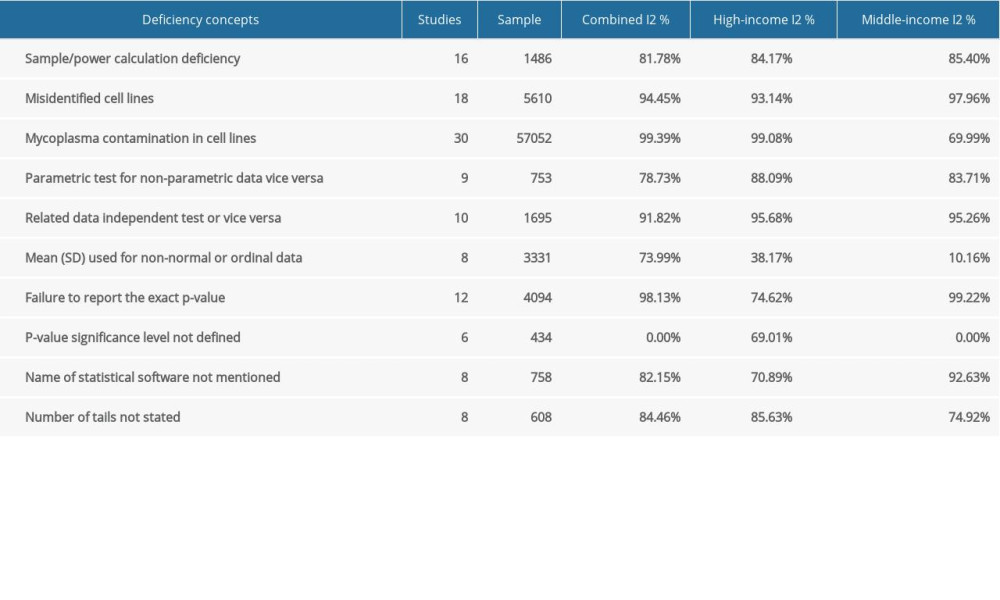 Table 2. Subgroup analysis of the estimated variation of reproducibility deficiencies in high-income and middle-income countries.
Table 2. Subgroup analysis of the estimated variation of reproducibility deficiencies in high-income and middle-income countries. Table 1. Baseline characteristics of the included studies.
Table 1. Baseline characteristics of the included studies. Table 2. Subgroup analysis of the estimated variation of reproducibility deficiencies in high-income and middle-income countries.
Table 2. Subgroup analysis of the estimated variation of reproducibility deficiencies in high-income and middle-income countries. In Press
15 Apr 2024 : Laboratory Research
The Role of Copper-Induced M2 Macrophage Polarization in Protecting Cartilage Matrix in OsteoarthritisMed Sci Monit In Press; DOI: 10.12659/MSM.943738
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
08 Mar 2024 : Animal Research
Modification of Experimental Model of Necrotizing Enterocolitis (NEC) in Rat Pups by Single Exposure to Hyp...Med Sci Monit In Press; DOI: 10.12659/MSM.943443
18 Apr 2024 : Clinical Research
Comparative Analysis of Open and Closed Sphincterotomy for the Treatment of Chronic Anal Fissure: Safety an...Med Sci Monit In Press; DOI: 10.12659/MSM.944127
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








