23 June 2022: Clinical Research
Expression of the Gene Polymorphism, SNP rs8192917, in 990 Han Chinese Patients with Postoperative Keloids
Xiulin Wen12CDE, Huicong Du1BF, Xiaoyan Hao3BF, Jingrong Wang4BF, Yuan Guo1AG*DOI: 10.12659/MSM.936963
Med Sci Monit 2022; 28:e936963
Abstract
BACKGROUND: A keloid is a pathological scar hyperplasia that is affected by genetic and environmental factors. Although the involvement of cytotoxic granzyme B in keloids has been recognized, there is almost no research on granzyme B (GZMB) gene polymorphisms and keloids. This study aimed to explore the relationship between genetic polymorphisms of GZMB and postsurgical keloid susceptibility in the Han Chinese population.
MATERIAL AND METHODS: A total of 3078 participants, including 990 patients with postsurgical keloids and 2088 controls without postsurgical keloids, were enrolled. We selected 15 common DNA variants in the GZMB gene for analysis. Associations were analyzed in both single marker-based and haplotype-based methods. The Genotype-Tissue Expression database was used to examine the biological significance of the targeted single nucleotide polymorphisms (SNPs).
RESULTS: SNP rs8192917 was found to be associated with the susceptibility of keloids (t statistic=4.82, P=1.47×10⁻⁶). An increased risk of keloids was significantly associated with the minor allele (C allele) of rs8192917 (odds ratio=1.33; 95% CI=1.18-1.49], P=1.47×10⁻⁶). In addition, a significant association was reported for genotypes of rs8192917 and clinical severity of keloids (χ²=10.61, P=0.03).
CONCLUSIONS: The results suggested there are significant associations between common genetic variants in GZMB and the susceptibility of postsurgical keloids in the Chinese Han population. These genetic polymorphisms were also related with the severity of postsurgical keloid symptoms in participants with keloids. The current study can contribute to future etiological and clinical research of keloids.
Keywords: Case-Control Studies, Disease Susceptibility, keloid, Polymorphism, Single Nucleotide, Alleles, China, Gene Frequency, Genetic Association Studies, Genetic Predisposition to Disease, Genotype, Granzymes, Humans, Postoperative Complications
Background
Keloid and hypertrophic scars represent an aberrant response to the wound healing process [1]. These scars are caused by cutaneous injuries, such as trauma, burns, surgeries, or uncertain origins, which is different from the origins of other scar types. Keloids are clinically treated as a benign skin tumor [2], and their incidence in the Asian population can be as high as 4% to 16% [3]. Keloid tissue is usually accompanied by chronic inflammation, and it is prone to infection and ulceration, which can lead to scar carcinoma [4,5]. Furthermore, for keloids, the prognosis of squamous cell carcinoma is poor and mortality is high [6]. Keloids can occur in locations including the ears, chin, chest, back, and vulva and are accompanied by unbearable itching and tingling and a serious negative impact on the quality of life and mental state of patients [7,8]. Due to the unclear pathogenesis of keloids, they have always posed a difficult problem in the field of plastic surgery.
There are currently many hypotheses about the formation of keloids, such as the immunology, tension, and endocrine hypotheses. Early studies have found that compared with in normal tissues, the concentrations of T lymphocytes, B lymphocytes, macrophages, and immune complexes in the peripheral circulation of keloids are significantly increased, and it is speculated that the proliferation of keloids may be caused by immune stimulation [9,10]. Genetic and molecular epidemiologic studies have suggested that the polymorphisms of many genes that affect the proliferation and apoptosis of keloid fibroblasts or extracellular deposition and degradation are associated with keloid risk, including TGF-β, Bcl-2, Smad7, IL-6, IL-4, IL-10, and Fas [11,12]. TGF-β is recognized as one of the most representative cytokines closely related to wound healing [13]. Given that hyperproliferation of fibroblasts is a major feature of keloids and that inflammation is associated with poor prognosis, the Fas/FasL pathway, which plays an important role in cell apoptosis, is thought to be associated with keloids. Some single nucleotide polymorphism (SNP) sites in exons 7 to 9 of the
The cytotoxic granzyme B (GzmB)/perforin pathway shows a similar function to the Fas/FasL pathway in apoptosis and inflammation, leading to poor healing of damaged tissues. There are 5 granzymes in humans, of which granzyme B is a serine protease encoded by the
To date, no study in Han Chinese individuals examining the association of the
Material and Methods
ETHICS STATEMENT:
Written informed consent was obtained from all participants. This study was approved by the Ethics Committee of the First Affiliated Hospital of Xi’an Jiaotong University per reference 20171016YG dated October 18, 2017, and the study procedures were performed following the tenets of the Declaration of Helsinki (version 2002).
STUDY PARTICIPANTS:
A total of 3078 study participants, including 990 patients with postsurgical keloids and 2088 control participants without postsurgical keloids, were consecutively recruited from the Plastic and Aesthetic Surgery Outpatient Department of the First Affiliated Hospital of Xi’an Jiaotong University between Jan 2018 and July 2021. All participants were unrelated Han Chinese individuals (at least 3 generations were of Han descent and had no history of migration). All participants were examined carefully by at least 2 dermatologists. According to the shape of the scars, proliferation outside the original wound boundary, and invasion of the surrounding normal skin, keloids were diagnosed. The diagnosis of the hypertrophic scar was confirmed when the height of the scar was more than 2 mm above the normal surrounding skin [25]. Patients with hypertrophic scars and some keloid syndromes were excluded from the study. The severity of keloids was assessed according to color, scar height, pliability, pain, and itching of keloid scars [26]. All healthy controls without keloids were from the same clinics, and all participants were free of obesity and autoimmune and systemic diseases in the study. Peripheral blood samples were collected from the study participants and preserved for further genotyping experiments. Demographic information, including age, sex, and family history of the study participants, was collected from questionnaires and medical records and is presented in Table 1.
SNP CHOICE AND EXPERIMENT FOR GENOTYPING:
GZMB is a short gene of only approximately 3.2 kb that includes untranslated regions. We extracted all common DNA variants (MAF ≥0.05) from the gene region based on data from the 1000 Genomes Project. We selected a total of 15 SNPs for genotyping (Supplementary Table 1). Genomic DNA was extracted using a commercial DNA kit (Axygen Scientific Inc, Union City, CA, USA). The candidate SNPs were genotyped based on the Sequenom MassARRAY platform, and the raw data were processed by a Typer Analyzer. Labels of samples (cases or controls) were blinded to technicians performing the genotyping experiments. Five percent of the study samples were replicated for genotyping experiments, and a 100% concordance rate was achieved.
STATISTICAL METHODS:
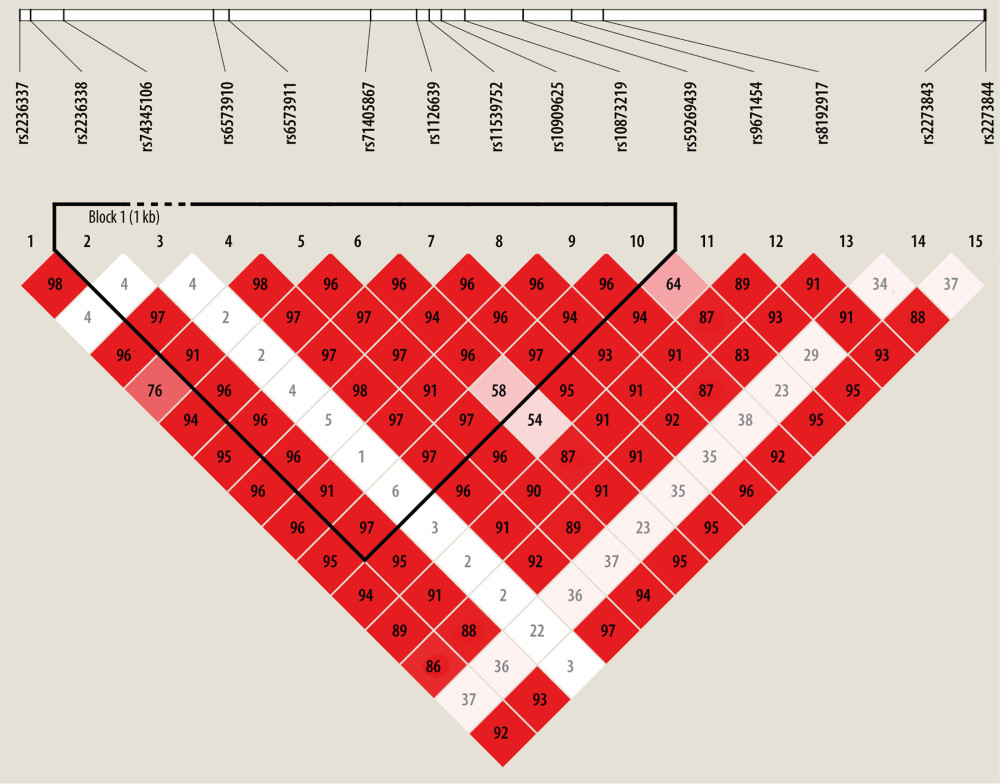
Power analysis was conducted using the GAS genetic power calculator (https://csg.sph.umich.edu/abecasis/gas_power_calculator/), and the result is summarized in Supplementary Figure 1. Characteristic and demographic variables were observed between participants with keloids and healthy controls. Genotyping quality was estimated by Hardy-Weinberg equilibrium tests conducted in the control group. Logit models were applied to each genetic marker for examining the association signals between genotypes and susceptibility to keloids. In the logistic model, the genotypes of each SNP were coded in additive mode. For 0, 1, or 2 copies of the minor allele, the SNP was coded as 0, 1, or 2. Plink [27] was utilized for fitting logistic models. Multiple testing was addressed by Bonferroni corrections through which the threshold P value was determined by 0.05 divided by the number of tests. Linkage disequilibrium (LD) patterns were analyzed using Haploview [28]. The standard method was applied to constructing LD blocks based on our genotyped SNPs [29]. Haplotypic frequencies in participants with keloids and controls were estimated and compared to identify significant haplotypes related to risk of keloids. In addition to the susceptibility to keloids, the relationship between genotypes of targeted SNPs and the clinical features of keloids were also investigated. The statistical computing program R was used for descriptive and general statistical analyses.
METHODS FOR IN SILICO ANALYSES:
In silico analyses were performed to investigate the biological significance of the top hit association signals identified in the association analyses. Several bioinformatics tools and databases were utilized. SIFT [30] and polyphen2 [31] were used to examine the biological significance of nonsynonymous variants on protein structures. The Genotype-Tissue Expression database was used to depict the potential expression quantitative trait loci (eQTL) pattern for candidate genes in various kinds of human tissues [32].
Results
DEMOGRAPHIC CHARACTERISTICS OF THE PARTICIPANTS:
With the formation of dermal hyalinized collagens visible under a microscope, keloids are seen to expand beyond the boundaries of the initial lesions. Hypertrophic scars, on the other hand, tend to form within the limits of wounds and present histologically as dermal nodules (Supplementary Figure 2). A total of 3078 participants, including 990 patients with keloids and 2088 healthy controls, were enrolled in this study. No significant differences were identified for age (t=−0.11, P=0.91), sex (χ2=0.0003, P=0.99), or family history (χ2=0.0857, P=0.77) between patients with keloids and healthy controls (Table 1). In the 990 patients with keloids, 273 patients (28%) had mild symptoms, 503 (51%) had moderate symptoms, and 214 (21%) had severe symptoms.
GENETIC ASSOCIATIONS BETWEEN SUSCEPTIBILITY TO KELOIDS AND GZMB GENETIC POLYMORPHISMS:
All of the 15 candidate SNPs were included in the Hardy-Weinberg equilibrium tests in the control group, and the results are summarized in Supplementary Table 1. SNP rs8192917 was found to be significantly related with the susceptibility of keloids (Table 2). An increased risk of keloids was associated with the C allele (minor allele) of rs8192917 (odds ratio [OR]=1.33; 95%CI=1.18–1.49, P=1.47×10−6). In addition to rs8192917, a couple of other SNPs achieved nominal significance despite their strong correlations with rs819291 (Figure 1). A large LD block comprising 9 SNPs was constructed (Figure 1). Significant associations were obtained from multiple haplotypes within these LD blocks (Table 3, χ2=329.60, P=2.81×10−67).
GENETIC ASSOCIATIONS FOR CLINICAL SEVERITY OF KELOIDS:
A significant association was observed for genotypes of rs8192917 and clinical severity of keloids (χ2=10.61, P=0.03). The proportion of patients with TT genotypes (homozygote of major alleles) that had mild and severe symptoms was 26% and 10%, respectively. On the other hand, the proportion of patients with CC genotypes (homozygote of minor alleles) that had mild and severe symptoms was 24% and 17%, respectively (Table 4). The proportion of patients with mild symptoms was lower for patients with TT genotypes than for patients with CC genotypes, while the proportion of patients with severe symptoms was higher for patients with TT genotypes than for patients with CC genotypes. The copy number of the C allele was associated with more severe symptoms in patients with keloids.
FUNCTIONAL CONSEQUENCES OF SNP RS8192917:
SNP rs8192917 is located in the exonic region of the GZMB gene. This SNP is a nonsynonymous variant, and its minor allele alters amino acids from Arg to Gln. Further bioinformatics tools were used to investigate whether this change in amino acids would in turn have significant functional consequences on proteins encoded by GZMB. The SIFT score was 0.189, and its prediction was “tolerated”. However, in Polyphen2, this SNP was predicted to be “possibly damaged”. Additionally, potential eQTL effects of this SNP on GZMB were also observed using the Genotype-Tissue Expression database. In tissue of the tibial nerve, a significant eQTL signal was observed (Supplementary Figure 3). The CC genotype of rs8192917 was significantly associated with a lower gene expression level of GZMB. Nevertheless, this eQTL signal was only identified in tissues of the tibial nerve, not in the other 45 kinds of human tissues, including the targeted tissue of keloids (Supplementary Table 2).
Discussion
Although multiple lines of evidence from studies of animal models have indicated that the
A previous study indicated that granzyme B (protein encoded by
The functional consequence of SNP rs8192917 was reported inconsistently in SIFT and Polyphen2. The SIFT score was 0.189, and its prediction was “tolerated”. However, in Polyphen2, this SNP was predicted to be “possibly damaged”. This is not very surprising, and a recent report focusing on
With the development of biotechnology, such as sequencing, genetic association analyses and multi-omics integrative analyses can help reveal the pathogenesis of complex diseases [36–39] and provide promising therapeutic candidates for the development of new drugs [40,41]. Therefore, sequencing-based integrative analysis might be a promising direction for determining the genetic architectures of
It is worth mentioning the limitations of the present study. Although all common DNA variants located within the gene region of
Conclusions
The results of this study suggest there are significant associations between the common genetic variants in the
Tables
Table 1. Characteristic and demographic information of the study participants.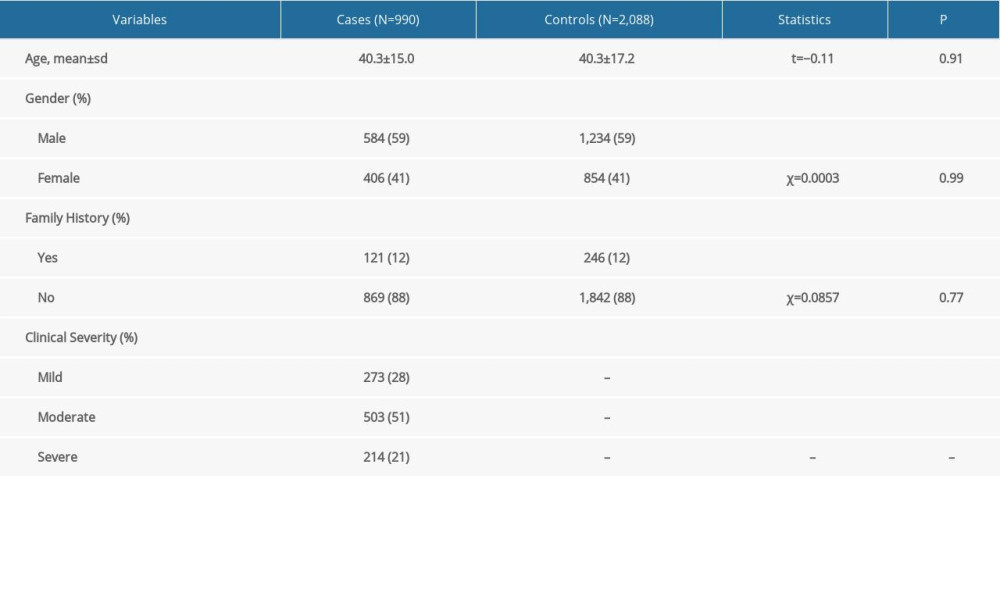 Table 2. Results of the single marker-based association analyses.
Table 2. Results of the single marker-based association analyses.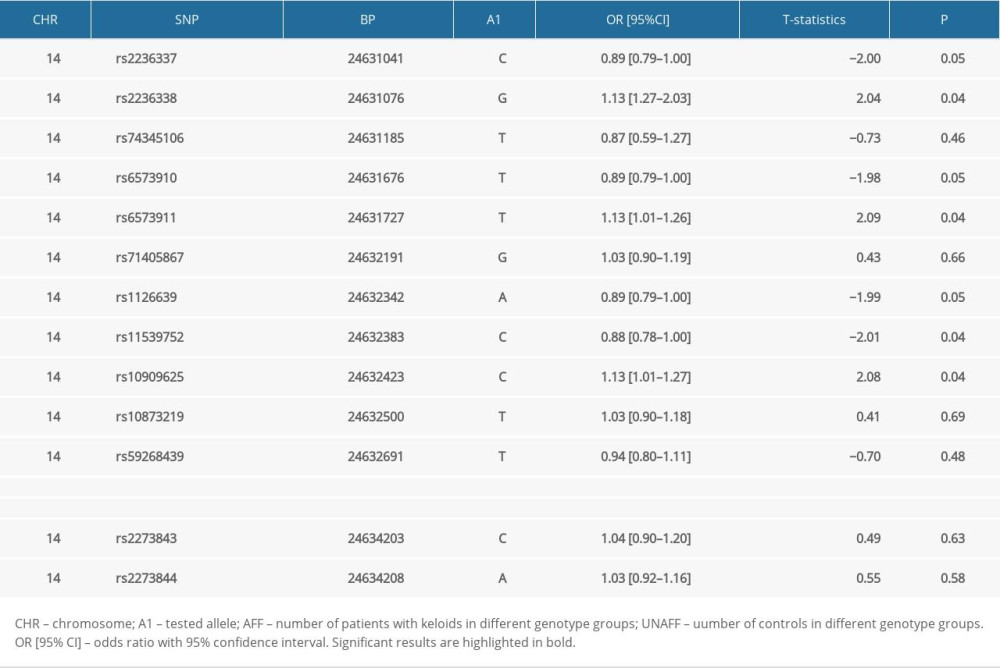 Table 3. Results of haplotype-based association analyses.
Table 3. Results of haplotype-based association analyses.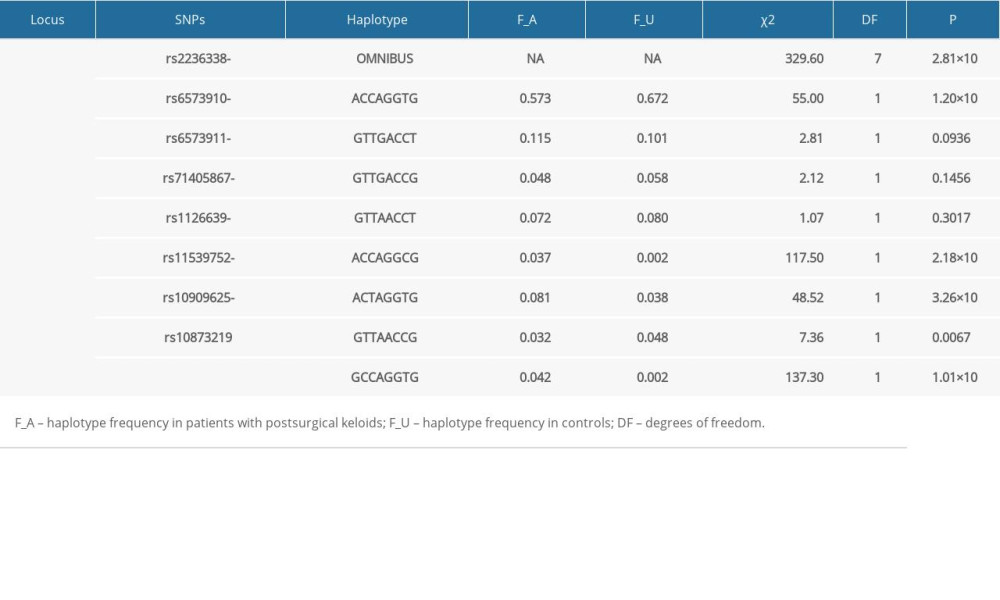 Table 4. Association between genotypes of rs8192917 and clinical severity of postsurgical keloids.
Table 4. Association between genotypes of rs8192917 and clinical severity of postsurgical keloids.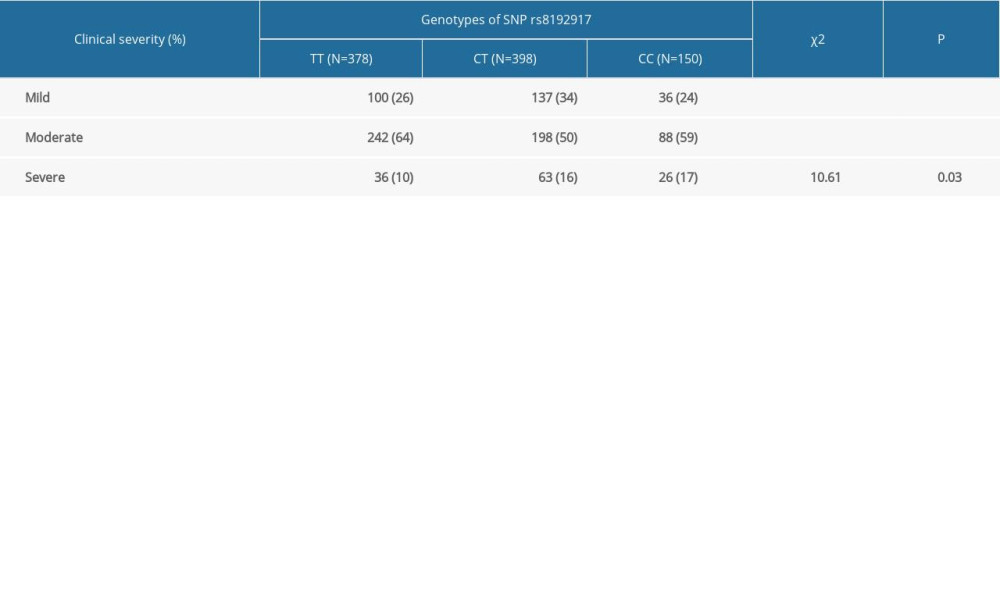 Supplementary Table 1. Basic information of the 15 genotyped SNPs.
Supplementary Table 1. Basic information of the 15 genotyped SNPs.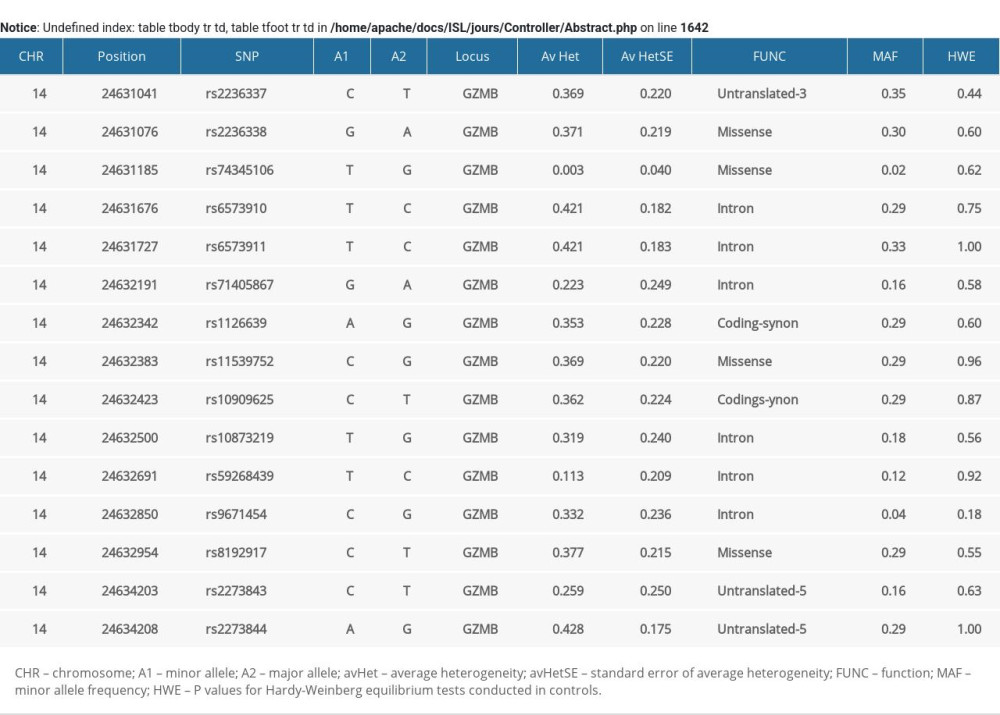 Supplementary Table 2. eQTL signals obtained for SNP rs8192917 on GZMB in 46 types of human tissues.
Supplementary Table 2. eQTL signals obtained for SNP rs8192917 on GZMB in 46 types of human tissues.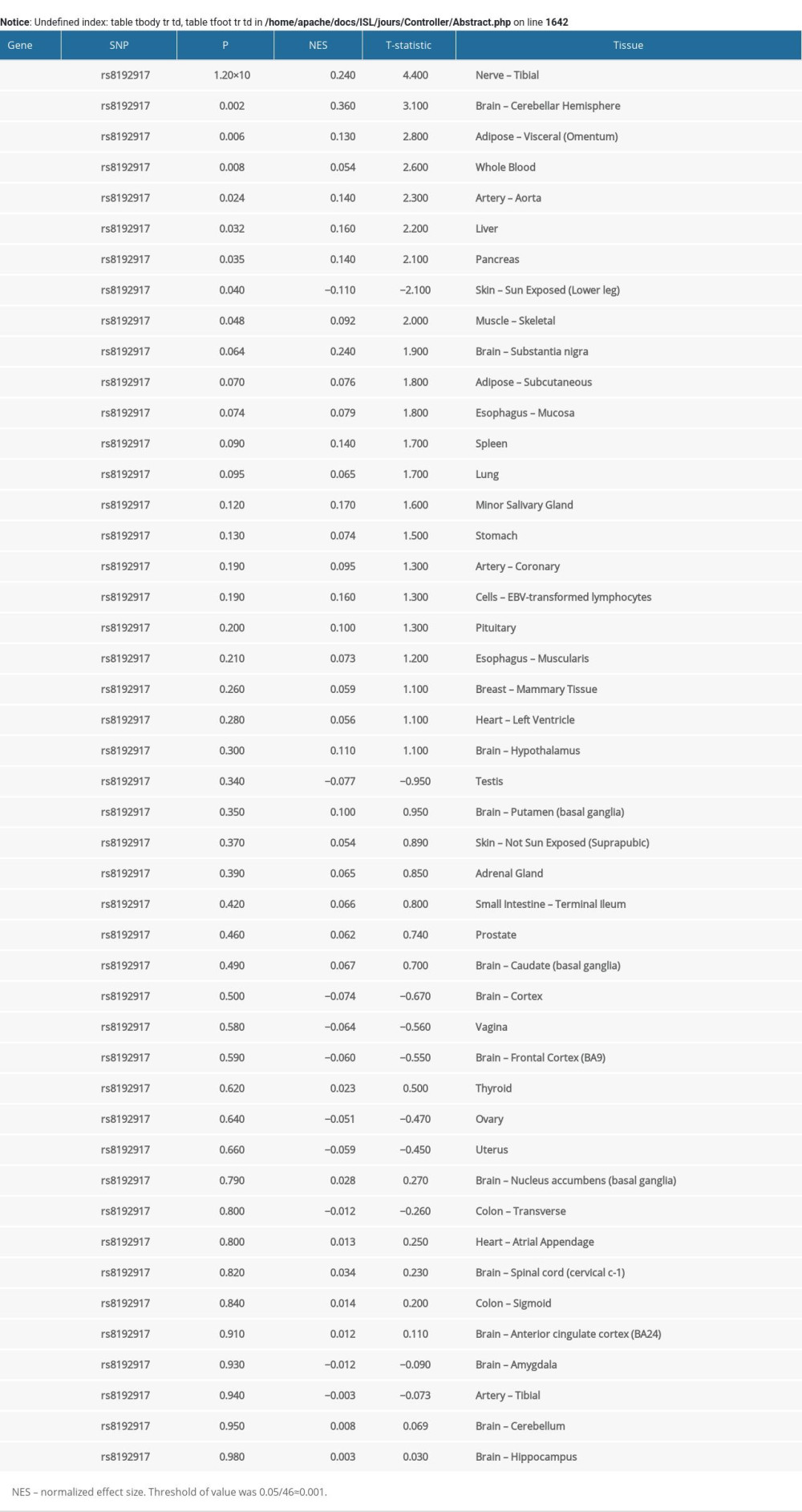
References
1. Berman B, Maderal A, Raphael B, Keloids and hypertrophic scars: Pathophysiology, classification, and treatment: Dermatol Surg, 2017(Suppl 1); S3-18
2. Viera MH, Caperton CV, Berman B, Advances in the treatment of keloids: J Drugs Dermatol, 2011; 10(5); 468-80
3. Satish L, Lyons-Weiler J, Hebda PA, Wells A, Gene expression patterns in isolated keloid fibroblasts: Wound Repair Regen, 2006; 14(4); 463-70
4. Trace AP, Enos CW, Mantel A, Harvey VM, Keloids and hypertrophic scars: A spectrum of clinical challenges: Am J Clin Dermatol, 2016; 17(3); 201-23
5. Ogawa R, Keloid and hypertrophic scars are the result of chronic inflammation in the reticular dermis: Int J Mol Sci, 2017; 18(3); 606
6. Majumder A, Srivastava S, Ranjan P, Squamous cell carcinoma arising in a keloid scar: Med J Armed Forces India, 2019; 75(2); 222-24
7. Kassi K, Kouame K, Kouassi A, Quality of life in black African patients with keloid scars: Dermatol Reports, 2020; 12(2); 8312
8. Olaitan PB, Keloids: Assessment of effects and psychosocial-impacts on subjects in a black African population: Indian J Dermatol Venereol Leprol, 2009; 75(4); 368-72
9. Murao N, Seino K, Hayashi T, Treg-enriched CD4+ T cells attenuate collagen synthesis in keloid fibroblasts: Exp Dermatol, 2014; 23(4); 266-71
10. Chen Z, Zhou L, Won T, Characterization of CD45RO(+) memory T lymphocytes in keloid disease: Br J Dermatol, 2018; 178(4); 940-50
11. Le QU, Chun-Di HEResearch advances on genetic and related gene in keloid: China Medical Abstract of Dermatology, 2015 [in Chinese]
12. Wang CH, Shan MJ, Liu H, Hyperbaric oxygen treatment on keloid tumor immune gene expression: Chin Med J (Engl), 2021; 134(18); 2205-13
13. Rhett JM, Ghatnekar GS, Palatinus JA, Novel therapies for scar reduction and regenerative healing of skin wounds: Trends Biotechnol, 2008; 26(4); 173-80
14. Liu X, Gao J, Li XExperimental study on fas gene death domain mutations in keloid pedigrees: Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi, 2007; 21(7); 698-701 [in Chinese]
15. Liu YB, Liu XJ, Gao JH, Duan HJAnalysis of the sequence of the variant exon-8 of fibroblastic Fas gene in keloid: Chinese Journal of Clinical Rehabilitation, 2006 [in Chinese]
16. Velotti F, Barchetta I, Cimini FA, Cavallo MG, Granzyme B in inflammatory diseases: Apoptosis, inflammation, extracellular matrix remodeling, epithelial-to-mesenchymal transition and fibrosis: Front Immunol, 2020; 11; 587581
17. Hiroyasu S, Hiroyasu A, Granville DJ, Tsuruta D, Pathological functions of granzyme B in inflammatory skin diseases: J Dermatol Sci, 2021; 104(2); 76-82
18. Carrino DA, Mesiano S, Barker NM, Proteoglycans of uterine fibroids and keloid scars: Similarity in their proteoglycan composition: Biochem J, 2012; 443; 361-68
19. Hiebert PR, Boivin WA, Abraham T, Granzyme B contributes to extracellular matrix remodeling and skin aging in apolipoprotein E knockout mice: Exp Gerontol, 2011; 46(6); 489-99
20. Hsu I, Parkinson LG, Shen Y, Serpina3n accelerates tissue repair in a diabetic mouse model of delayed wound healing: Cell Death Dis, 2014; 5; e1458
21. Heusel JW, Wesselschmidt RL, Shresta S, Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells: Cell, 1994; 76(6); 977-87
22. Turner CT, Bolsoni J, Zeglinski MR, Granzyme B mediates impaired healing of pressure injuries in aged skin: NPJ Aging Mech Dis, 2021; 7(1); 6
23. Zhu F, Wu B, Li P, Association study confirmed susceptibility loci with keloid in the Chinese Han population: PLoS One, 2013; 8(5); e62377
24. Bakry OA, Younes SF, Abdel Wahed M, Evaluation of the role of granzyme B in exuberant scar pathogenesis: An immunohistochemical study: Anal Quant Cytopathol Histpathol, 2015; 37(4); 221-26
25. Baryza M, Baryza G, The Vancouver Scar Scale: An administration tool and its interrater reliability: J Burn Care Rehabil, 1995; 16; 535-38
26. Idriss N, Maibach H, Scar assessment scales: A dermatologic overview: Skin Res Technol, 2009; 15(1); 1-5
27. Purcell S, Neale B, Todd-Brown K, PLINK: A tool set for whole-genome association and population-based linkage analyses: Am J Hum Genet, 2007; 81(3); 559-75
28. Barrett JC, Fry B, Maller J, Daly MJ, Haploview: Analysis and visualization of LD and haplotype maps: Bioinformatics, 2005; 21(2); 263-65
29. Gabriel SB, Schaffner SF, Nguyen H, The structure of haplotype blocks in the human genome: Science, 2002; 296(5576); 2225-29
30. Vaser R, Adusumalli S, Leng SN, SIFT missense predictions for genomes: Nat Protoc, 2016; 11(1); 1-9
31. Adzhubei IA, Schmidt S, Peshkin L, A method and server for predicting damaging missense mutations: Nat Methods, 2010; 7(4); 248-49
32. Consortium GT, The Genotype-Tissue Expression (GTEx) project: Nat Genet, 2013; 45(6); 580-85
33. Jin Y, Birlea SA, Fain PR, Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo: N Engl J Med, 2010; 362(18); 1686-97
34. Jin Y, Andersen G, Yorgov D, Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants: Nat Genet, 2016; 48(11); 1418-24
35. Ernst C, Hahnen E, Engel C, Performance of in silico prediction tools for the classification of rare BRCA1/2 missense variants in clinical diagnostics: BMC Med Genomics, 2018; 11(1); 35
36. Guan F, Zhang T, Han W, Relationship of SNAP25 variants with schizophrenia and antipsychotic-induced weight change in large-scale schizophrenia patients: Schizophr Res, 2020; 215; 250-55
37. Zhang TX, Zhu L, Ni T, Voltage-gated calcium channel activity and complex related genes and schizophrenia: A systematic investigation based on Han Chinese population: J Psychiatr Res, 2018; 106; 99-105
38. Han W, Zhang TX, Ni T, Relationship of common variants in CHRNA5 with early-onset schizophrenia and executive function: Schizophr Res, 2018; 206; 407-12
39. Guan F, Ni T, Han W, Evaluation of the relationships of the WBP1L gene with schizophrenia and the general psychopathology scale based on a case-control study: Am J Med Genet B Neuropsychiatr Genet, 2020; 183; 164-71
40. Guan F, Ni T, Zhu W, Integrative omics of schizophrenia: From genetic determinants to clinical classification and risk prediction: Mol Psychiatry, 2021; 188; 1-14
41. Shen C, Li H, Li M, DLRAPom: A hybrid pipeline of Optimized XGBoost-guided integrative multiomics analysis for identifying targetable disease-related lncRNA-miRNA-mRNA regulatory axes: Brief Bioinform, 2022; 23(2); bbac046
Tables
 Table 1. Characteristic and demographic information of the study participants.
Table 1. Characteristic and demographic information of the study participants. Table 2. Results of the single marker-based association analyses.
Table 2. Results of the single marker-based association analyses. Table 3. Results of haplotype-based association analyses.
Table 3. Results of haplotype-based association analyses. Table 4. Association between genotypes of rs8192917 and clinical severity of postsurgical keloids.
Table 4. Association between genotypes of rs8192917 and clinical severity of postsurgical keloids. Table 1. Characteristic and demographic information of the study participants.
Table 1. Characteristic and demographic information of the study participants. Table 2. Results of the single marker-based association analyses.
Table 2. Results of the single marker-based association analyses. Table 3. Results of haplotype-based association analyses.
Table 3. Results of haplotype-based association analyses. Table 4. Association between genotypes of rs8192917 and clinical severity of postsurgical keloids.
Table 4. Association between genotypes of rs8192917 and clinical severity of postsurgical keloids. Supplementary Table 1. Basic information of the 15 genotyped SNPs.
Supplementary Table 1. Basic information of the 15 genotyped SNPs. Supplementary Table 2. eQTL signals obtained for SNP rs8192917 on GZMB in 46 types of human tissues.
Supplementary Table 2. eQTL signals obtained for SNP rs8192917 on GZMB in 46 types of human tissues. In Press
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952