27 July 2020: Laboratory Research
Biochemical Feature of LMO2 Interactome and LMO2 Function Prospect
Wenhao Wang1BC, Yaxin Chen1BC, Ying Chang2AD, Wei Sun1EFG*DOI: 10.12659/MSMBR.924421
Med Sci Monit Basic Res 2020; 26:e924421
Abstract
BACKGROUND: LMO2 belongs to the LIM-Only group of LIM domain protein superfamily. It is ubiquitously expressed in different types of tissues and locates either in the nucleus or in the cytoplasm depending on the tissue type. Till now the unique function of LMO2 was considered to be serving as a bridging or blocking molecule that mediates extensive protein-protein interactions. However, the exactly biological features of LMO2 interactome as well as LMO2 function spectrum remain largely unclear.
MATERIAL AND METHODS: In this study, yeast 2-hybrid assay was firstly performed using LMO2 as the bait and the characteristic of LMO2 protein interactome was analyzed according to the yeast 2-hybrid data and other relative biological information primarily using bioinformatic method.
RESULTS: Our data indicated that LMO2 favored interacting with peptides containing β-sheet structure and having relatively unstable confirmation. Moreover, several LMO2 favored interacting domains were identified, including WD40 repeat, coiled-coil, Ankyrin repeat, Zinc finger, PDZ, and SH3, and functions of these domain-containing members were dramatically enriched in some types of cancers.
CONCLUSIONS: Our results revealed a LMO2 favored protein-interaction pattern in both secondary structure and domain level, and concentrated LMO2 function in kinds of cytoplasmic metabolism pathways as well as multiple types of cancers.
Keywords: Databases, Nucleic Acid, Molecular Biology, Two-Hybrid System Techniques, Adaptor Proteins, Signal Transducing, LIM Domain Proteins, Peptides, Protein Binding, Protein Domains, Protein Interaction Mapping, Proto-Oncogene Proteins
Background
The human
Material and Methods
YEAST 2-HYBRID (Y2H) ASSAY:
The assay was performed with Matchmaker™ Gold Yeast 2-Hybrid system (Clontech, Palo Alto, CA, USA). In brief, full-length LMO2 fusing with GAL4 DNA binding domain (GAD4-BD) was constructed as the bait and the pre-transformed human universal cDNA library from Clontech was used for screening. Mating and selecting procedures were stringently following the instruction from the manufacturer. Finally, positive clones were picked out, used as the template for polymerase chain reaction (PCR) amplification and the amplified fragments from each clone were sequenced.
BIOINFORMATIC ANALYSIS METHODS:
All sequences from Y2H positive clones were subjected to both Nucleotide and Protein BLAST (Basic Local Alignment Search Tool) on NCBI website (
Information of subcellular distribution of core LMO2 interactome members and the extended gene list related to the recurrent LMO2-interacted domains were retrieved from Genecards Database (https://www.genecards.org/). The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment assays were performed by R ClusterProfiler package [17] with default parameters. Relative plots were drawn by R ggplot2 package.
Results
LMO2 HAS A WIDE SPECTRUM OF INTERACTION MEMBERS WITH SEVERAL PREFERRED DOMAINS AND PEPTIDE FEATURES:
A quadruple selection system (−Ade/−His/+X-a-Gal/+Aureobasidin A) was used for the Y2H screening (Supplementary Figure 1A). Toxicity and autoactivation of GAL4-BD-LMO2 construct were firstly excluded by normal yeast growth on SD/-TRP plate whilst completely growth inhibition on SD/−TRP/X/A plate, and the known LMO2-LDB1 interaction as the positive control was confirmed by positive yeast growth on SD/−Leu/−TRP/X/A (DDO/X/A) plate and blue colonies (Supplementary Figure 1B). After mating of the GAL4-BD-LMO2 strain with GAL4-AD library, positive clones were firstly selected on the DDO/X/A plates for the first round screening and then blue colonies were re-seeded on the QDO/X/A plates (SD/−Ade/−His/−Leu/−Trp/+X-a-Gal/+Aureobasidin A) for the second round (stringent) screening (Supplementary Figure 1C). The finally positive clones after stringent screening were PCR amplified and sequenced, and a total of 205 sequences as the potentially LMO2 interacting partners were obtained (Supplementary Table 1). DNA and protein blast showed that among all these sequences, 123 were matched with the coding sequence (ORF) region of certain genes, whilst 50 in 3′-UTR, 4 in 5′-UTR, 7 in intron, 3 in LncRNA, 11 in intergenic region, and 8 that could not match any region of human genome (NA), all of which represented a false-positive part of the Y2H result (Figure 1A). Additionally, CD search of the 123 coding sequences identified 103 sequences containing conserved domains, and some typical domains, including WD40 repeat, coiled-coil, Ankyrin repeat, Zinc finger, FN3, SH3, metallo-dependent hydrolases, and NADB_Rossmann, recurrently appeared in these members (Figure 1B, Supplementary Table 1). These domains represented a repertoire of LMO2 favored interacting domains and of note, although PDZ domain appeared only once, our previous work had demonstrated that LMO2 could bind with the PDZ domain of disheveled proteins [4], indicating that PDZ domain should be an authentic interacting partner of LMO2 as well.
The in frame BD-fusion peptides from non-coding sequences, although considered as the artificial section of the Y2H screening, together with the rest of the coding sequences without any matched conserved domains, might provide additional biochemical and biophysical features of LMO2 interactome. Analysis on such set of peptides revealed that the peptides length ranged from 7aa to 275aa, and LMO2 favored the peptides containing α-helix or β-sheet secondary structure or both, particularly the β-sheet, with the exception of just 5 members containing only random coils (Figure 1C). Notably, LMO2 also favored to bind to the peptides with relatively unstable confirmation (Figure 1C, above the threshold), balanced hydrophility/hydrophobity (Figure 1D, most members concentrated at −0.5 – +0.5 of average hydropathicity value) but had no preference on isoelectric point (Figure 1D, pI ranged from 3.79 to 12.1).
FUNCTION PROSPECT OF LMO2 VIA ITS CORE INTERACTING PARTNERS AND EXTENDED RELATED GENES:
Although LMO2 has been reported to distribute either in nucleus or cytoplasm, the subcellular distribution of its core interactome members showed a wide spectrum, including nucleus, cytosol, mitochondrion, membrane system as well as extracellular, and these members covered a large cellular function categories (Figure 2A, 2B, and Supplementary Table 2). Moreover, according to the public data from Genecards Database (https://www.genecards.org/), we found that most of these members had a dominant but generally more than one distinct subcellular localization, with a majority had been functional annotated by GO or KEGG whilst a minority not yet (function unclear, Figure 2A, 2B). KEGG and GO (biological process [BP] and cellular component [CC]) functional enrichment assay revealed that the core LMO2 partners primarily enriched on the functions of carbohydrate metabolism, particularly glucose, propanoate, and lactate metabolism; nucleoside metabolism, including both purine and pyrimidine metabolism; co-enzyme metabolism; ribosome and mitochondrion related functions (Figure 2C), altogether implementing the potentially cytosol function complexity of LMO2.
Since the LMO2 preferred binding domains might exist in more members other than those Y2H assay identified and we might therefore miss some information, we next collected the extended structural and functional related genes of 6 primary domains, including WD40 repeat, coiled-coil, Ankyrin repeat, Zinc finger, PDZ, and SH3 (gene list available on Genecards Database, https://www.genecards.org/) and performed additional functional enrichment assay. Notably, related genes from each of the 6 domains along, in the union set or intersect of the 6 domains were all dramatically enriched in relations with a majority types of cancers and several classic cancer related functions, particularly for the 6 domain intersect genes which represented a more concentrated and confident set of LMO2 functional related members (Figure 2D). Altogether these results strongly suggested LMO2 a tightly cancer-related gene.
Discussion
The LIM domain is well studied as a characteristic module that mediates multiple protein-protein interactions [5]. LMO2, due to its LIM domain-only structural feature, lacks any enzymatic or DNA binding activities and is generally considered as a “bridging or blocking” molecule. Although short peptide length (only 158 aa) and simple functional domains, LMO2 has rather pivotal functions. Early studies showed that
In this study, we also identified several LMO2 favored interacting domains, including WD40 repeat, coiled-coil, Ankyrin repeat, Zinc finger, PDZ, and SH3. However, there was a defect that some known LMO2 binding partners, such as LDB1, GATA1, TAL1, and LYL1, were failed to be screened out by Y2H. This was possibly due to the composition bias of the screening library and/or sequencing failure of some positive clones, and implied that some positive LMO2 interaction partners might be missed. To address this issue, we expanded the LMO2 interaction repertoire by domain similarities. All the 6 domain containing protein families have multiple members and their functional related genes, all of which represent the extended repertoire of LMO2 functional targeted candidates. Notably, these genes were distinctively enriched in the cancer related functions, indicating that the predominant functions of LMO2 are involved in cancers. Previous literatures indicated that nuclear LMO2 could either activate or inhibit gene transcription depending on interaction with different partners and binding to different DNA motifs [8–10,12]. As a cytoplasmic protein, LMO2 also functions as either an oncogene or a tumor suppressor via interaction with a variety of proteins and participates in multiple cellular processes [4,14,15,26]. The structural feature of LMO2, as well as the diversity of its interaction spectrum, suggests that it may be consumed by different partners and thus involved in multiple cellular pathways simultaneously in a certain cell type, and the interaction preference of LMO2, as well as the relative abundance ratio of LMO2 and other partners, may altogether determine the diversified predominant functions of LMO2 in different cell backgrounds.
Conclusions
Taken together, this study depicted an overview of LMO2 favored protein-interaction pattern in both secondary structure and domain level, and concentrated LMO2 function in some of cytosol metabolism pathways as well as multiple types of cancers.
Supplementary Data
Figures
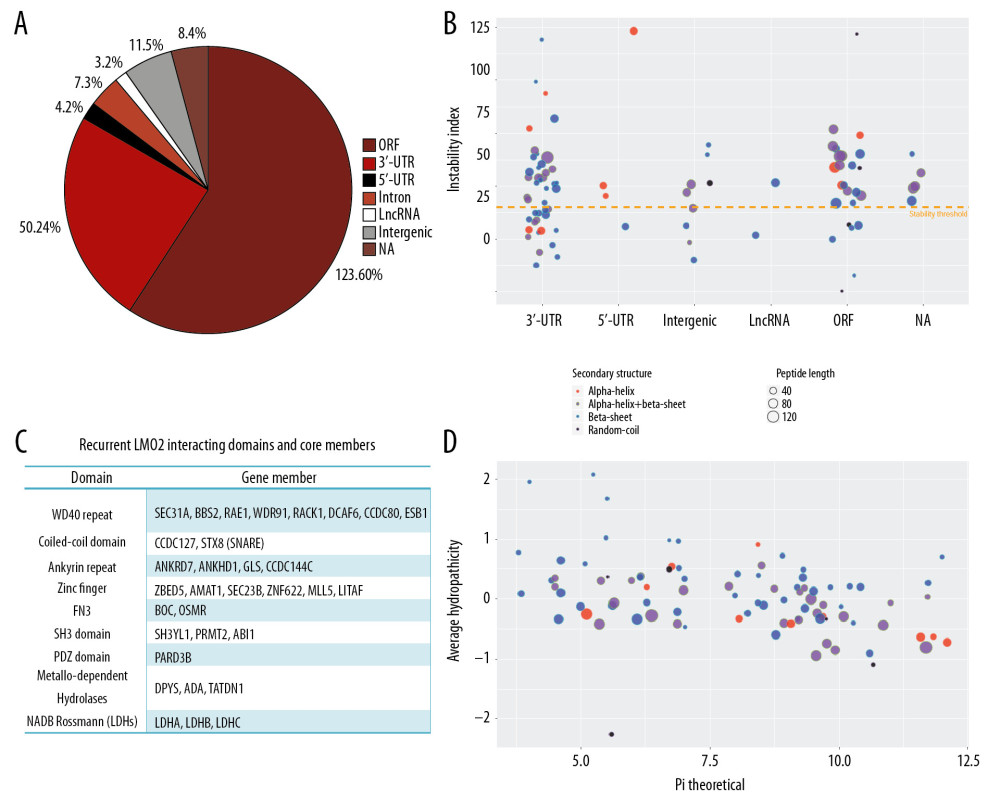 Figure 1. LMO2 has a wide spectrum of interaction members with several preferred domains and peptide features. (A) Pie plot showing the distribution of all positive LMO2-binding sequences from Y2H screening in each category. (B) The list of recurrently appeared domains and their relevant members in LMO2 core interactome. (C) The bubble plot showing the distribution of length, predicted secondary structure and stability of LMO2 binding peptides without any conserved domains in each category. Relative information was marked on the plot. (D) The bubble plot showing the distribution of predicted hydropathicity and isoelectric point (pI) of LMO2 binding peptides without any conserved domains. Relative information was marked on the plot. Y2H – yeast 2-hybrid; ORF – open reading frame; 3′-UTR – 3′-untranslated region; 5′-UTR – 5′-untranslated region; lncRNA – long non-coding RNA; NA – not available.
Figure 1. LMO2 has a wide spectrum of interaction members with several preferred domains and peptide features. (A) Pie plot showing the distribution of all positive LMO2-binding sequences from Y2H screening in each category. (B) The list of recurrently appeared domains and their relevant members in LMO2 core interactome. (C) The bubble plot showing the distribution of length, predicted secondary structure and stability of LMO2 binding peptides without any conserved domains in each category. Relative information was marked on the plot. (D) The bubble plot showing the distribution of predicted hydropathicity and isoelectric point (pI) of LMO2 binding peptides without any conserved domains. Relative information was marked on the plot. Y2H – yeast 2-hybrid; ORF – open reading frame; 3′-UTR – 3′-untranslated region; 5′-UTR – 5′-untranslated region; lncRNA – long non-coding RNA; NA – not available. 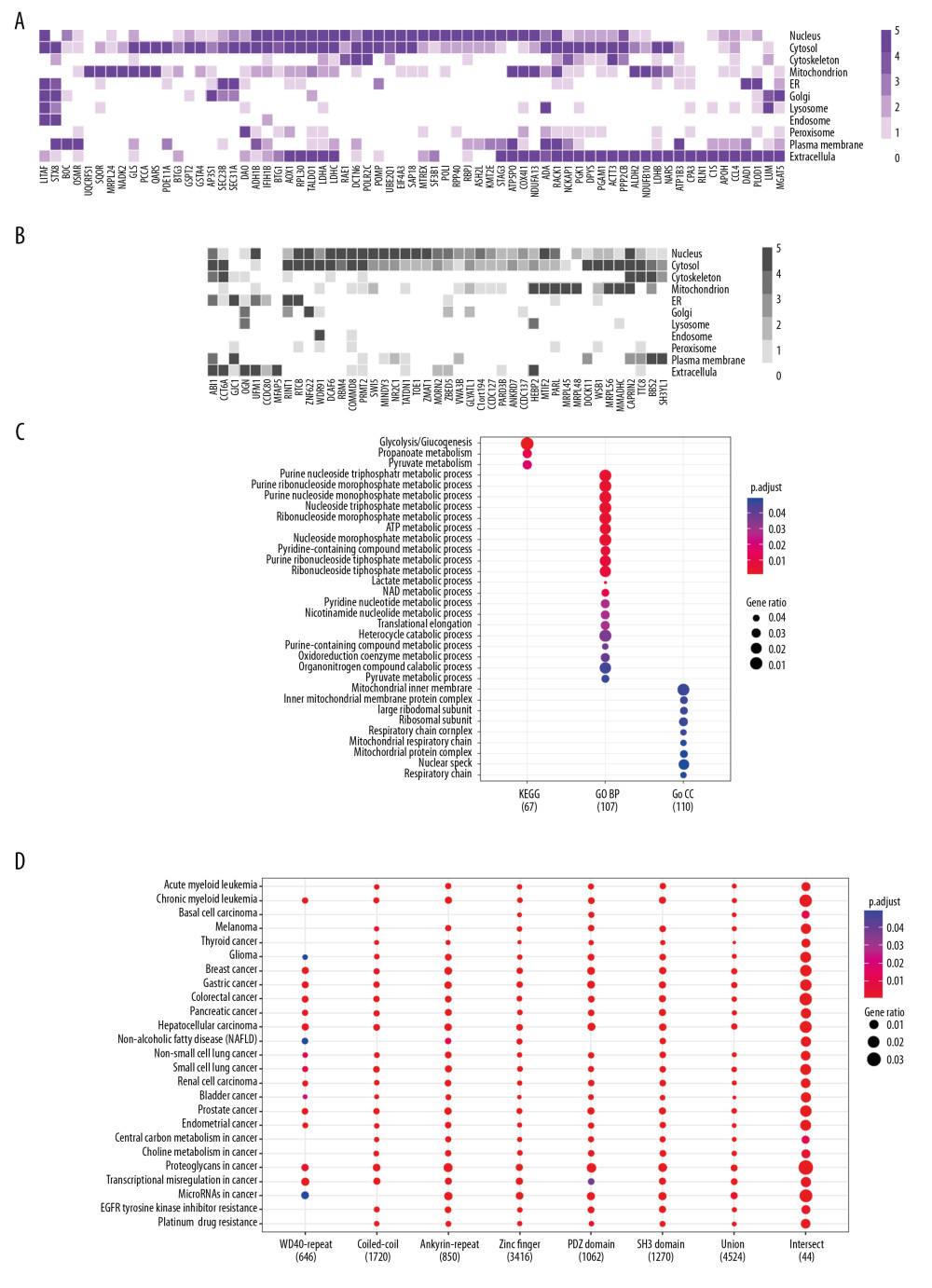 Figure 2. Function prospect of LMO2 via its core interacting partners and extended related genes. (A) Heatmap showing the subcellular distribution of LMO2 core interactome members with KEGG or GO annotation and (B) Heatmap showing the subcellular distribution of LMO2 core interactome members without KEGG or GO annotation. Legend from 5 to 0 indicated the normalized subcellular abundance from highest to none. (C) Dot plot showing the enriched KEGG, GO_BP and GO_CC terms from the enrichment assay on LMO2 core interactome members. Relative information was marked on the plot. (D) Dot plot showing the enriched cancer related KEGG terms from the enrichment assay on extended 6 domain related members. Relative information was marked on the plot. GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; BP – biological process; CC – cellular component.
Figure 2. Function prospect of LMO2 via its core interacting partners and extended related genes. (A) Heatmap showing the subcellular distribution of LMO2 core interactome members with KEGG or GO annotation and (B) Heatmap showing the subcellular distribution of LMO2 core interactome members without KEGG or GO annotation. Legend from 5 to 0 indicated the normalized subcellular abundance from highest to none. (C) Dot plot showing the enriched KEGG, GO_BP and GO_CC terms from the enrichment assay on LMO2 core interactome members. Relative information was marked on the plot. (D) Dot plot showing the enriched cancer related KEGG terms from the enrichment assay on extended 6 domain related members. Relative information was marked on the plot. GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; BP – biological process; CC – cellular component. References
1. Boehm T, Foroni L, Kaneko Y, The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13: Proc Natl Acad Sci USA, 1991; 88; 4367-71
2. Royer-Pokora B, Loos U, Ludwig WD, TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11): Oncogene, 1991; 6; 1887-93
3. Agostinelli C, Paterson JC, Gupta R, Detection of LIM domain only 2 (LMO2) in normal human tissues and haematopoietic and non-haematopoietic tumours using a newly developed rabbit monoclonal antibody: Histopathology, 2012; 61; 33-46
4. Liu Y, Huang D, Wang YZ, LMO2 attenuates tumor growth by targeting the Wnt signaling pathway in breast and colorectal cancer: Sci Rep, 2016; 6; 36050
5. Dawid IB, Breen JJ, Toyama R, Lim domains: Multiple roles as adapters and functional modifiers in protein interactions: Trends Genet, 1998; 14; 156-62
6. McCormack MP, Rabbitts TH, Activation of the T-cell oncogene LMO2 after gene therapy for X-linked severe combined immunodeficiency: N Engl J Med, 2004; 350; 913-22
7. Wadman IA, Osada H, Grutz GG, The LIM-only protein LMO2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins: EMBO J, 1997; 16; 3145-57
8. Grutz GG, Bucher K, Lavenir I, The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells: EMBO J, 1998; 17; 4594-605
9. Sun W, Yang S, Shen WW, Identification of DeltaEF1 as a novel target that is negatively regulated by LMO2 in T-cell leukemia: Eur J Haematol, 2010; 85; 508-19
10. Sun W, Shen WW, Yang S, Homo-binding character of LMO2 isoforms and their synergic and antagonistic functions in regulating hematopoietic-related target genes: J Biomed Sci, 2010; 17; 22
11. Fleskens V, Mokry M, van der Leun AM, FOXP3 can modulate TAL1 transcriptional activity through interaction with LMO2: Oncogene, 2016; 35; 4141-48
12. Chambers J, Rabbitts TH, LMO2 at 25 years: a paradigm of chromosomal translocation proteins: Open Biol, 2015; 5; 150062
13. Cubedo E, Gentles AJ, Huang C, Identification of LMO2 transcriptome and interactome in diffuse large B-cell lymphoma: Blood, 2012; 119; 5478-91
14. Liu Y, Wang ZY, Huang D, LMO2 promotes tumor cell invasion and metastasis in basal-type breast cancer by altering actin cytoskeleton remodeling: Oncotarget, 2017; 8; 9513-24
15. Liu Y, Wu C, Zhu TH, LMO2 enhances lamellipodia/filopodia formation in basal-type breast cancer cells by mediating ARP3-profilin1 interaction: Med Sci Monit, 2017; 23; 695-703
16. Wu C, Liu Y, Gu XX, LMO2 blocks the UBA6-USE1 interaction and downstream FAT10ylation by targeting the ubiquitin fold domain of UBA6: Biochem Biophys Res Commun, 2016; 478; 1442-48
17. Yu G, Wang L, Han Y, clusterProfiler: An R package for comparing biological themes among gene clusters: OMICS, 2012; 16; 284-87
18. Warren AJ, College WH, Carlton MB, The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development: Cell, 1994; 78; 45-57
19. Yamada Y, Pannell R, Forster A, Rabbitts TH, The oncogenic LIM-only transcription factor Lmo2 regulates angiogenesis but not vasculogenesis in mice: Proc Natl Acad Sci USA, 2000; 97(1); 320-24
20. Larson RC, Fisch P, Larson TA, T cell tumours of disparate phenotype in mice transgenic for Rbtn-2: Oncogene, 1994; 9(12); 3675-81
21. Hacein-Bey-Abina S, Von Kalle C, Schmidt M, LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1 [published correction appears in Science, 2003; 302(5645): 568]: Science, 2003; 302(5644); 415-19
22. Pike-Overzet K, de Ridder D, Weerkamp F, Ectopic retroviral expression of LMO2, but not IL2Rgamma, blocks human T-cell development from CD34+ cells: Implications for leukemogenesis in gene therapy: Leukemia, 2007; 21(4); 754-63
23. McCormack MP, Young LF, Vasudevan S, The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal: Science, 2010; 327(5967); 879-83
24. Dastmalchi S, Wilkinson-White L, Kwan AH, Solution structure of a tethered Lmo2(LIM2)/Ldb1(LID) complex: Protein Sci, 2012; 21(11); 1768-74
25. Sewell H, Tanaka T, El Omari K, Conformational flexibility of the oncogenic protein LMO2 primes the formation of the multi-protein transcription complex: Sci Rep, 2014; 4; 3643
26. Liu Y, Yuan M, Wu C, A comprehensive function analysis of LMO2 in different breast cancer subtypes: Oncotarget, 2017; 9(10); 8911-26
Figures
 Figure 1. LMO2 has a wide spectrum of interaction members with several preferred domains and peptide features. (A) Pie plot showing the distribution of all positive LMO2-binding sequences from Y2H screening in each category. (B) The list of recurrently appeared domains and their relevant members in LMO2 core interactome. (C) The bubble plot showing the distribution of length, predicted secondary structure and stability of LMO2 binding peptides without any conserved domains in each category. Relative information was marked on the plot. (D) The bubble plot showing the distribution of predicted hydropathicity and isoelectric point (pI) of LMO2 binding peptides without any conserved domains. Relative information was marked on the plot. Y2H – yeast 2-hybrid; ORF – open reading frame; 3′-UTR – 3′-untranslated region; 5′-UTR – 5′-untranslated region; lncRNA – long non-coding RNA; NA – not available.
Figure 1. LMO2 has a wide spectrum of interaction members with several preferred domains and peptide features. (A) Pie plot showing the distribution of all positive LMO2-binding sequences from Y2H screening in each category. (B) The list of recurrently appeared domains and their relevant members in LMO2 core interactome. (C) The bubble plot showing the distribution of length, predicted secondary structure and stability of LMO2 binding peptides without any conserved domains in each category. Relative information was marked on the plot. (D) The bubble plot showing the distribution of predicted hydropathicity and isoelectric point (pI) of LMO2 binding peptides without any conserved domains. Relative information was marked on the plot. Y2H – yeast 2-hybrid; ORF – open reading frame; 3′-UTR – 3′-untranslated region; 5′-UTR – 5′-untranslated region; lncRNA – long non-coding RNA; NA – not available. Figure 2. Function prospect of LMO2 via its core interacting partners and extended related genes. (A) Heatmap showing the subcellular distribution of LMO2 core interactome members with KEGG or GO annotation and (B) Heatmap showing the subcellular distribution of LMO2 core interactome members without KEGG or GO annotation. Legend from 5 to 0 indicated the normalized subcellular abundance from highest to none. (C) Dot plot showing the enriched KEGG, GO_BP and GO_CC terms from the enrichment assay on LMO2 core interactome members. Relative information was marked on the plot. (D) Dot plot showing the enriched cancer related KEGG terms from the enrichment assay on extended 6 domain related members. Relative information was marked on the plot. GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; BP – biological process; CC – cellular component.
Figure 2. Function prospect of LMO2 via its core interacting partners and extended related genes. (A) Heatmap showing the subcellular distribution of LMO2 core interactome members with KEGG or GO annotation and (B) Heatmap showing the subcellular distribution of LMO2 core interactome members without KEGG or GO annotation. Legend from 5 to 0 indicated the normalized subcellular abundance from highest to none. (C) Dot plot showing the enriched KEGG, GO_BP and GO_CC terms from the enrichment assay on LMO2 core interactome members. Relative information was marked on the plot. (D) Dot plot showing the enriched cancer related KEGG terms from the enrichment assay on extended 6 domain related members. Relative information was marked on the plot. GO – Gene Ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes; BP – biological process; CC – cellular component. Most Viewed Current Articles
15 Jun 2022 : Clinical Research
Evaluation of Apical Leakage After Root Canal Obturation with Glass Ionomer, Resin, and Zinc Oxide Eugenol ...DOI :10.12659/MSMBR.936675
Med Sci Monit Basic Res 2022; 28:e936675
07 Jul 2022 : Laboratory Research
Cytotoxicity, Apoptosis, Migration Inhibition, and Autophagy-Induced by Crude Ricin from Ricinus communis S...DOI :10.12659/MSMBR.936683
Med Sci Monit Basic Res 2022; 28:e936683
01 Jun 2022 : Laboratory Research
Comparison of Sealing Abilities Among Zinc Oxide Eugenol Root-Canal Filling Cement, Antibacterial Biocerami...DOI :10.12659/MSMBR.936319
Med Sci Monit Basic Res 2022; 28:e936319
08 Dec 2022 : Original article
Use of Estimated Glomerular Filtration Rate and Urine Albumin-to-Creatinine Ratio Based on KDIGO 2012 Guide...DOI :10.12659/MSMBR.938176
Med Sci Monit Basic Res 2022; 28:e938176









