05 November 2020: Lab/In Vitro Research
Integrated Bioinformatics Analysis of Hub Genes and Pathways Associated with a Compression Model of Spinal Cord Injury in Rats
Chang Jiang1BCE, Zheng Li1BCE, Zhaoyi Wu1DEF, Yun Liang1CD, Lixia Jin1BD, Yuanwu Cao1CG, Shengcheng Wan1DG, Zixian Chen1AG*DOI: 10.12659/MSM.927107
Med Sci Monit 2020; 26:e927107
Abstract
BACKGROUND: Spinal cord injury (SCI) is a serious nervous system condition that can cause lifelong disability. The aim of this study was to identify potential molecular mechanisms and therapeutic targets for SCI.
MATERIAL AND METHODS: We constructed a weighted gene coexpression network and predicted which hub genes are involved in SCI. A compression model of SCI was established in 45 Sprague-Dawley rats, which were divided into 5 groups (n=9 per group): a sham operation group, and 1, 3, 5, and 7 days post-SCI groups. The spinal cord tissue on the injured site was harvested on 1, 3, 5, and 7 days after SCI and 3 days after surgery in the sham operation group. High-throughput sequencing was applied to investigate the expression profile of the mRNA in all samples. Differentially expressed genes were screened and included in weighted gene coexpression network analysis (WGCNA). Co-expressed modules and hub genes were identified by WGCNA. The biological functions of each module were investigated using the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes databases.
RESULTS: According to the RNA-seq data, a total of 1965 differentially expressed genes were screened, and WGCNA identified 10 coexpression modules and 5 hub genes. Module function analysis revealed that SCI was associated with immune response, cell division, neuron projection development, and collagen fibril organization.
CONCLUSIONS: Our study revealed dynamic changes in a variety of biological processes following SCI and identified 5 hub genes via WGCNA. These results provide insights into the molecular mechanisms and therapeutic targets of SCI.
Keywords: Genetic Association Studies, pathologic processes, Spinal Cord Injuries, Computational Biology, Gene Expression Profiling, Gene Expression Regulation, gene ontology, Gene Regulatory Networks, Spinal Cord Compression
Background
Traumatic spinal cord injury (SCI) is an important medical condition that causes disability in patients and imposes a heavy burden on patients’ families and the economy. A recent study estimated that there were 930 000 new cases of SCI worldwide, with an age-standardized incidence rate of 13 per 100 000 and 9.5 million years of life lived with disability. Ominously, SCI affects mostly young adults [1]. Compared with other central nervous system injuries, such as brain injury, SCI can result in greater long-term burden because patients with SCI usually survive the injury but have a poor life quality.
The pathophysiology of acute SCI involves primary (mechanical) and secondary mechanisms. Although both of these factors cause spinal cord damage and loss of motion, the secondary mechanisms warrant greater attention because they involve excessive inflammation, lipid peroxidation, inhibition of neurite growth, and delayed cell loss, which may be preventable or treatable [2,3]. Therefore, it is important to identify the key biological processes and hub genes involved in the secondary mechanisms of SCI.
High-throughput sequencing technology has become widely used in the field of SCI research. Transcriptome sequencing helped us identify many vital differentially expressed genes by comparing post-SCI and control samples.
Many molecular mechanisms were revealed by the functional analysis and in-depth bioinformatics analysis of these genes. However, the wealth of these abundant data, if only for differential expression analysis, was often underused, reducing the value of high-throughput screening. Weighted gene coexpression network analysis (WGCNA) is a systematic biology approach that converts coexpression measures into linkage weight or overlapping topological measures and is widely used to explore the interactions between gene networks [4,5]. Generally, genes involved in the same pathway or functional complex show similar expression patterns and can be regarded as a module. WGCNA can be used to detect modules of highly correlated genes, summarize these clusters using the module eigengene or an intramodular hub gene, and correlate modules with other modules and external sample traits [6]. WGCNA is very suitable for monitoring the overall changes of hundreds of genes, in order to explore their relationships and the occurrence and development of disease, greatly increasing the availability of sequencing data. Therefore, it is beneficial to apply WGCNA to the study of pathological mechanisms of secondary SCI.
Most spinal cord injuries in the clinic feature a compressive component, while, at present, there has been no report of WGCNA being applied in the RNA sequencing result of spinal cord compressive injury. In this study, we used a compressive SCI model and sequenced the mRNAs of spinal cord samples at different times post-SCI. According to the WGCNA results, 10 co-expressed modules and 5 hub genes were identified, which may help to illustrate the molecular mechanisms of SCI and identify promising therapeutic targets.
Material and Methods
ANIMALS AND MODEL OF COMPRESSIVE SCI:
A total of 45 male Sprague-Dawley rats (body weight 200 g to 250 g) were used in this study. The rats were obtained from Shanghai SLAC Laboratory Animal Co., Ltd. All rats were raised in a specific pathogen-free barrier system with a standard 12–12 h light/dark cycle at 23±1°C with free access to food and water in the laboratory animal center of Zhongshan Hospital, Shanghai, China. The animals were allowed to acclimate to the new system for 1 week before surgery was performed. All animals were anesthetized by intraperitoneal injection of 5% chloral hydrate (0.6 mL/100 g body weight). During the operation, an electric blanket was used to maintain the body temperature of the rats, and ketoprofen (subcutaneous, 5 mg per kg body weight) was used for pain control. The rats were placed in independent cages after surgery. Each animal received penicillin by intramuscular injection (20 000 UI per 100 g body weight in 0.5 mL sterile saline) in the first 3 postoperative days to reduce infection. The animals’ bladders were expressed using the Credes maneuver (abdominopelvic compression) every 12 h to deal with SCI-related urinary retention until the rats were euthanized. This study was approved by the Committee of Ethics on Animal Experiments of Zhongshan Hospital.
The rats were randomly divided into 5 groups (n=9 per group): a sham operation group, and 1, 3, 5, and 7 days post-SCI groups. Within each group, 3 rats were used for RNA-seq and the others for histological examination (half for coronal section and half for transverse section). To prepare the clip compression model of SCI, the rats were anesthetized and underwent laminectomy at T8–9. Compressive injury was induced in the SCI groups by applying a 30 g aneurysm clip for 1 min, as previously described [7,8]. Rats in the sham operation group underwent laminectomy to expose the spinal cord, without clipping.
The animals in the SCI groups were euthanized at the corresponding time points, while the sham group was euthanized on the third postoperative day. The rats were euthanized by deep anesthetization via inhalation of isoflurane (4%). Tissue samples were subsequently quickly harvested, and the rats eventually died from the irreversible blood loss from tissue sampling.
HISTOLOGICAL EXAMINATION:
The spinal cords of the upper and lower 10 mm centered on the injured site were collected immediately and fixed in 4% paraformaldehyde, dehydrated in gradient alcohol, cleared in xylene, and embedded in paraffin. Then the embedded tissues were cut into 5-μm thick slices. The coronal section contained the full length of the longitudinal axis of the tissues, passing through the center of the spinal cord, and the transverse section was located at the site of the injury. These sections were then stained with hematoxylin and eosin (H&E) (Servicebio, Wuhan, China). The histological changes after SCI were recorded and analyzed under an optical imaging system (Leica Microsystems, Bensheim, Germany).
RNA ISOLATION AND SEQUENCING:
Spinal cord tissue from 5 mm around the epicenter of damage was harvested at the indicated times after inducing SCI and stored in liquid nitrogen. Total RNA was extracted using TRI Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The RNA concentration and quality were assessed using an automated electrophoresis system (Agilent 2100 Bioanalyzer, Agilent Technologies, Santa Clara, CA, USA). Subsequently, mRNA was purified from total RNA using oligo (dT)-attached magnetic beads for generating a cDNA library. The cDNA fragments were then sequenced on a sequencer (BGISEQ-500, Beijing Genomics Institute, Shenzhen, China). After quality control, raw data were mapped to the reference genome RAT6.0 and gene expression analysis was performed.
IDENTIFICATION OF DIFFERENTIALLY EXPRESSED GENES AND WGCNA:
Differentially expressed genes (DEGs) in the post-SCI groups were identified as genes that showed an increase in expression by >2 fold or a decrease by <0.5 fold relative to the sham group using the ‘limma’ package (v3.24.15) in R v3.4.1 [9]. The adjusted P values obtained by applying the Benjamini-Hochberg method for multiple testing correction (false discovery rate [FDR]) were considered significant when ≤0.05 [10]. Any post-SCI group with reads per kilobase per million (FPKM) >1 was used to construct the coexpressing modules. The gene coexpression network was constructed using the WGCNA package (v1.66) in R, as previously described [4,6]. All DEGs were included in the analysis. Briefly, the ‘pickSoftThreshold’ function was used to determine the correct power-β after considering the smallest value to yield a free-scale topology. We selected 16 as the power value to obtain modules containing highly coexpressing genes. Using a dynamic tree-cutting algorithm and merging threshold function at 0.25, we identified 10 modules and summarized the expression profile of each module by its eigengenes. Expression trends of each module were plotted using the average fold change of the top 100 eigengenes, relative to the sham group, at each time point to reflect the overall change trend of each module.
IDENTIFICATION AND VISUALIZATION OF HUB GENES:
We combined modules with similar expression trends to create new sets of genes. The top 150 connections among hub genes in each set were visualized into a weighted coexpression network using Cytoscape (v3.4.0) (http://www.cytoscape.org) [11]. Intramodular hub genes are usually located in the center of the module. The thickness of the edge was based on ‘weight’, which referred to the connection strength between 2 nodes (genes) in terms of the correlation value obtained from the ‘TOMsimilarity’ function in WGCNA. The Topological overlap matrix (TOM), which was obtained from ‘TOMsimilarity’, gave these values for the respective genes. This TOM value is always between 0 and 1. A higher value refers to a strong connection or coexpression of genes.
GENE ENRICHMENT ANALYSIS:
The Gene Ontology (GO) [12,13] and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.kegg.jp/) databases were used for functional annotation of specific modules using the Database for Annotation, Visualization and Integrated Discovery (DAVID) [14,15]. FDR <0.05 was considered as the threshold to indicate a statistically significant difference in the analysis.
Results
MORPHOLOGICAL CHANGES OF SCI AFTER COMPRESSION:
The animal model of SCI was established as shown in Figure 1A. All 45 rats received successful operations, with hindlimb paralysis and urinary retention observed in SCI rats and no obvious abnormalities observed in the sham group. Spinal cord disruption was visible after spinal cord compression (Figure 1B–1G). Images of the spinal cord and H&E-stained tissue sections revealed the extent of the pathological changes to the spinal cord (Figure 1H–1Q). Blood cells were visible 1 day after the injury, and tissue sections contained numerous red blood cells along the injury site with abundant inflammatory cells. Loss of the surrounding nerve tissue due to secondary injury was visible on day 3, at which time the blood had been absorbed. The tissue had undergone severe collapse, and scarring was evident. By day 5, the scar had grown in size with ongoing tissue repair, and the inflammation had diminished alongside the formation of a spinal cord cavity. By day 7, the transverse diameter of the spinal cord had decreased without obvious signs of clamp gross injury, but the injury site was mostly filled with scar tissue and a cavity.
SCREENING OF DEGS:
A total of 15 samples of spinal cord tissues from the injured rats (including 3 samples from the sham group) were homogenized and sequenced by BGISEQ-500 (Beijing Genomics Institute, China). Each group comprised 3 replicates. We searched for genes that were significantly upregulated or downregulated at any time compared with the sham group, as shown in Table 1, and identified a total of 1965 DEGs (Figure 2A–2D) for inclusion in the WGCNA.
WGCNA:
We performed WGCNA to develop gene modules related to the pathology of SCI. For this screening, we set β=16 and identified the genes using the dynamicTreeCut algorithm in R. A total of 10 modules were split and each module was assigned a different color (Table 2). Heat maps were plotted to characterize the expression of these 10 modules. In this analysis, genes in the magenta and brown modules were downregulated on day 1 after injury, and tended to recover after this time. Genes in the blue, pink, green, grey, and red modules were downregulated on day 1 after injury, and then started to recover on day 3 and kept increasing after that. Generally, black and turquoise modules decreased, while the black module had a sudden decline on day 1 and then recovered. The expression of genes in the yellow module was upregulated on day 3, plateauing thereafter. The expression trends of genes in each module are shown in Figure 3A–3T.
BIOLOGICAL FUNCTIONS ANALYSIS:
We used DAVID to determine the biological functions of the genes in the 10 modules in terms of GO biological process terms and KEGG pathways. Table 2 provides an overview of the 10 modules, which contained the top 3 biological processes (BP) identified by GO analysis. Four of these modules were focused on the results of their KEGG pathway analysis, as illustrated in Figure 3U–3X. Genes in the largest module, turquoise, were related to chemical synaptic transmission, sensory perception of pain, and neurotransmitter secretion. These genes kept decreasing with time, especially at 5 days post-SCI, indicating they may have been regulated by injury-related factors, hindering neuronal remodeling. Genes in the blue module were related to immune function, being enriched with genes involved in inflammatory responses, innate immune responses, and adaptive immune responses. Pathway analysis also indicated that they were mainly related to inflammatory response. The blue module showed transient downregulation after injury followed by a gradual increase in expression, indicating that these genes may be involved in secondary damage, and that the immune system might play important roles in secondary injury and repair of the spinal cord following compressive injury. The GO-BP and KEGG analyses of the brown module indicated it was related to cell division, chromosome segregation, and mitotic nuclear division. These genes were markedly downregulated following injury but tended to recover after the acute stage. The green module showed a relationship with the extracellular matrix, collagen fibrils, and blood vessels according to GO-BP and KEGG analyses. The genes in the green module were suppressed on the third day, and then gradually increased and exceeded the baseline, which suggested a process of tissue reconstruction or glial scar formation.
IDENTIFICATION OF HUB GENES:
The modules with similar expression trends were firstly merged to obtain a larger gene network to help avoid the loss of marginal genes in some small modules, such as pink, black, and magenta. According to the expression results shown in Figure 3A, we merged the modules into 4 sets: set A (blue+green+grey+pink+red), set B (brown+magenta), set C (turquoise+black), and set D (yellow). Network analysis of the 4 sets allowed us to identify the driver genes via centralized hub analysis. We found that set A comprised 3 subsets, in which cytochrome P-450 (CYP) 2D1 (Cyp2d1) was the hub gene, while C2 and Cyth4 were the key nodes linking the subsets. For the other 3 sets, we found that the hub genes were Bub1b and Tpx2 for set B, sodium-potassium ATPase (Atp1a3) for set C, and Rrp9 and Pgam5 for set D (Figure 4). These hub genes were selected based on their degree of connection with other genes in the WGCNA network. The other major hub genes are listed in Table 3.
Discussion
RNA sequencing has been used extensively to identify potential therapeutic targets and prognostic markers in SCI. The pathological mechanism of SCI is complex and involves more than 20 potential pathological processes. To date, most studies that have performed RNA-seq in SCI have not fully exploited the data collected. Differential analysis and function enrichment analysis were commonly used for mining RNA-seq data, but not enough for the massive data of high-throughput sequencing. In this study, we used WGCNA for a deeper analysis of RNA-seq; it integrated 1965 DEGs into 10 modules. Four relatively explainable modules were identified through the function enrichment analysis. They were the turquoise module related to pain or sense perception, the brown module related to cell division, the blue module related to immune response, and the green module related to extracellular matrix organization. By integrating similar modules into 4 gene sets, we identified
Inflammation is a double-edged sword for neurons in SCI. It is essential for the local tissue repairing but toxic if remained unresolved, which results in regeneration disorder [16]. In the present study, the increased expression of
The reconstruction of neural pathways is essential during the functional recovery after SCI.
Glial scarring is a healing response but is unfavorable to axon growth in the long term [27]. Extracellular matrix organization is considered an important feature of scar formation [28]. In the present study, the green module showed a relationship with extracellular matrix, collagen fibril, and blood vessel development that suggested a process of tissue reconstruction, or glial scar formation. It was suppressed initially after SCI, and then gradually increased even beyond the baseline. This suggested that extracellular matrix failed to be activated in the early stage and failed to limit the expansion of inflammation, but was improperly activated in the later stage, causing obstruction of neural regeneration. This finding suggested that the precise regulation of extracellular matrix organization may be a potential therapeutic target for spinal cord injury.
Cell proliferation is a manifestation of tissue remodeling. The main pathways included in the brown module, such as cell division, chromosome segregation, and mitotic nuclear division, embodied this feature. We also identified
WGCNA is an effective method for analyzing biological systems. In the present study, WGCNA found many hub genes and pathways that would not be noticed by routine differential analysis. A small number of studies on SCI applied WGCNA. For example, Wang et al. used WGCNA in 2 open access array data and identified 3 hub genes including
This study had the following limitations. First, the spinal cord is a complex tissue with many cell types, and sequencing reflects expression changes in the whole tissue, rather than in specific cells. This may mistakenly attribute some functions to certain cells. More in-depth research in to related types of cells is needed. The emerging topographic single-cell sequencing technology may solve this problem but it is too expensive and not mature in spinal cord tissue. Second, we did not validate the changes in gene expression in terms of protein expression. Due to the post-transcriptional regulation, the trend of protein expression may be different from that of mRNA. Third, we obtained tissue samples up to 7 days after SCI, so future studies should include long-term observation following SCI. Finally, we failed to establish a hypothesis on the function of the last hub gene,
Conclusions
In this study, we constructed a weighted gene coexpression network and predicted hub genes and essential biological processes in compressive spinal cord injury (SCI). Four essential modules were revealed to be associated with chemical synaptic transmission, immune response, cell division, and extracellular matrix organization. Moreover,
Figures
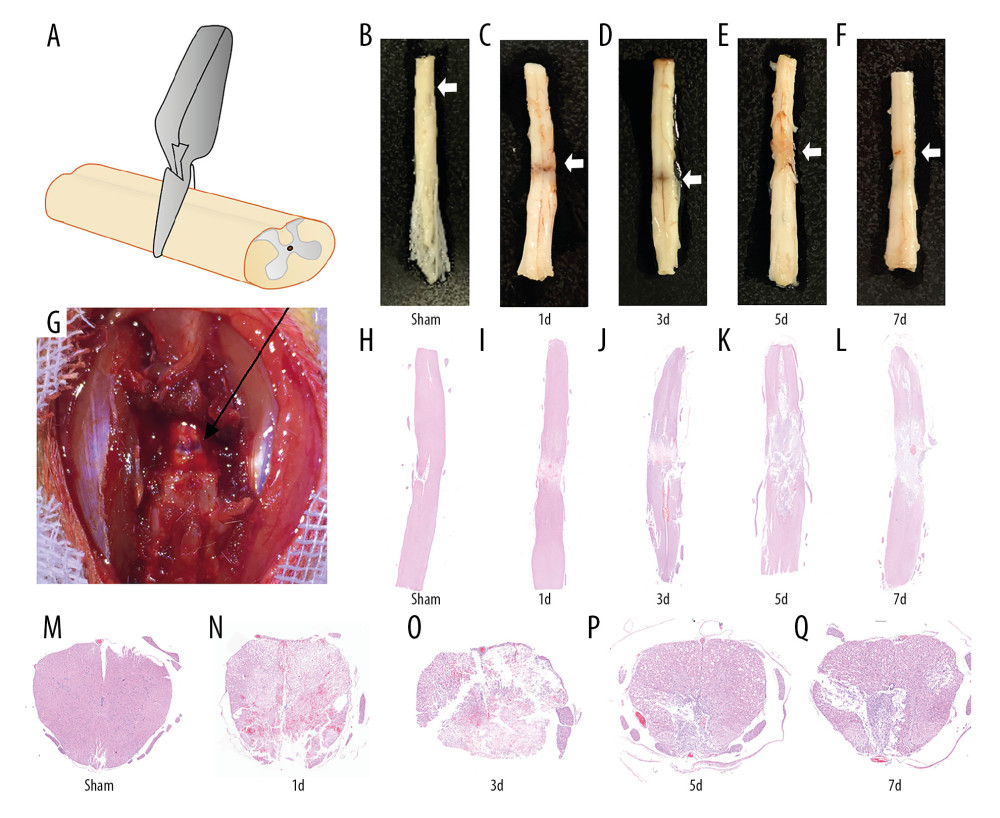 Figure 1. Establishment of spinal cord injury after compression. (A) Diagram of compression model of spinal cord injury (SCI). (B–G) The figure demonstrated the visible signs of bleeding and spinal cord disruption after SCI. (H–Q) H&E staining showed the pathological changes at different time points (Sham, 1d, 3d, 5d, and 7d) after SCI in, coronal, and transverse sections (white arrows mark the injury site).
Figure 1. Establishment of spinal cord injury after compression. (A) Diagram of compression model of spinal cord injury (SCI). (B–G) The figure demonstrated the visible signs of bleeding and spinal cord disruption after SCI. (H–Q) H&E staining showed the pathological changes at different time points (Sham, 1d, 3d, 5d, and 7d) after SCI in, coronal, and transverse sections (white arrows mark the injury site). 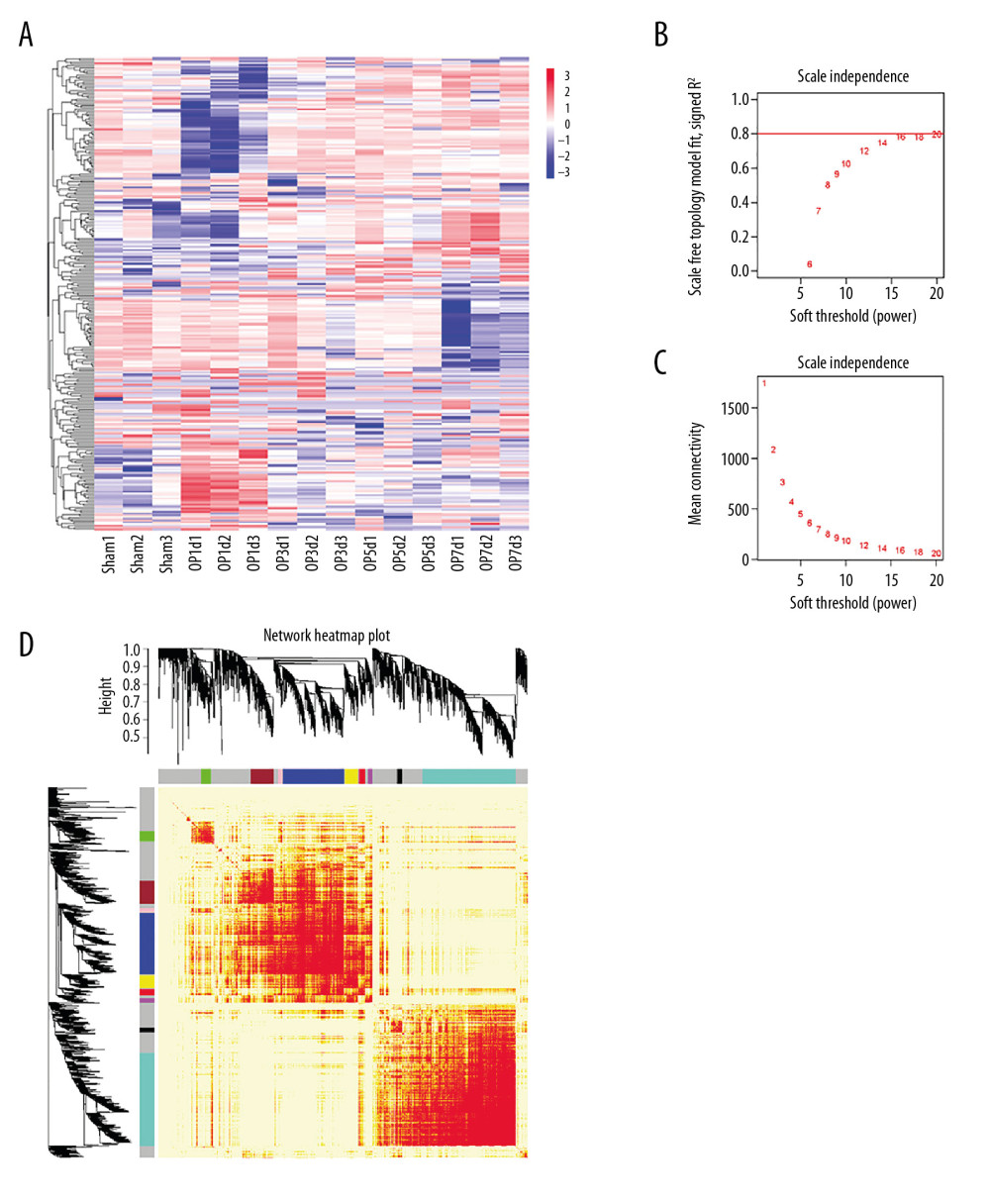 Figure 2. Differential gene screening and weighted gene coexpression network analysis. (A) A total of 1965 differential genes were screened at different time points (Sham, 1d, 3d, 5d, and 7d) after spinal cord injury (SCI). (B, C) Analysis of network topology showed that it met the scale-free topology threshold of 0.8 when β=16. (D) The heatmap of topological matrix showed the correlation network, in which each color represented a gene module.
Figure 2. Differential gene screening and weighted gene coexpression network analysis. (A) A total of 1965 differential genes were screened at different time points (Sham, 1d, 3d, 5d, and 7d) after spinal cord injury (SCI). (B, C) Analysis of network topology showed that it met the scale-free topology threshold of 0.8 when β=16. (D) The heatmap of topological matrix showed the correlation network, in which each color represented a gene module. 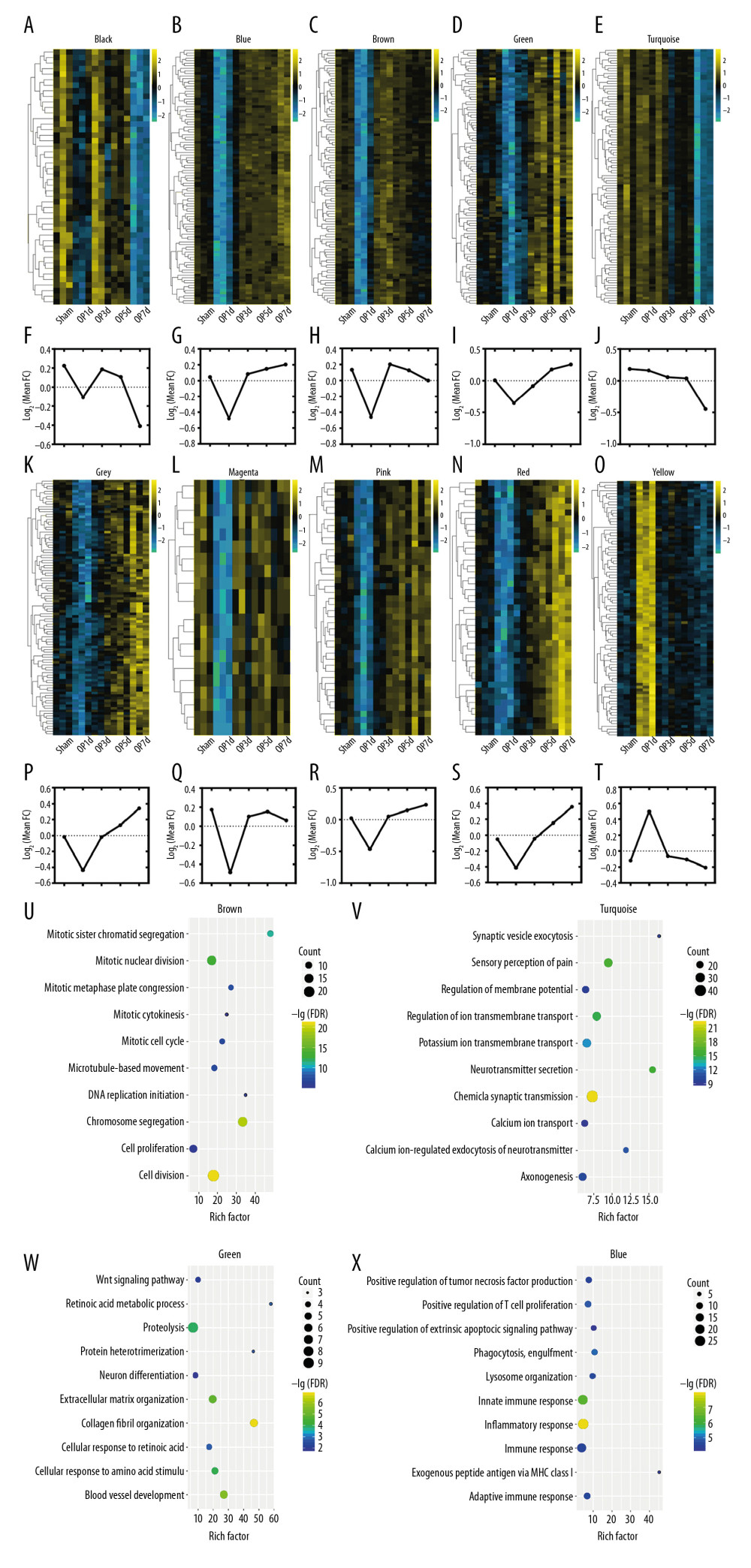 Figure 3. Expression trends and pathway enrichment of modules identified via WGCNA. (A–T) A total of 10 modules were split and each module was assigned a symbolic color. It was found that the modules of magenta and brown were sharply downregulated on the first day after injury, then tended to recover; the modules of blue, pink, green, grey, and red were downregulated after injury, and then rebounded upward 3 days after SCI; the modules of black and turquoise showed a continuous decrease after injury; the yellow module was sharply upregulated within 3 days after injury, and then tended to be flat. (U–X) The figures demonstrated the top 10 of pathway enrichment in blue, brown, green, and turquoise.
Figure 3. Expression trends and pathway enrichment of modules identified via WGCNA. (A–T) A total of 10 modules were split and each module was assigned a symbolic color. It was found that the modules of magenta and brown were sharply downregulated on the first day after injury, then tended to recover; the modules of blue, pink, green, grey, and red were downregulated after injury, and then rebounded upward 3 days after SCI; the modules of black and turquoise showed a continuous decrease after injury; the yellow module was sharply upregulated within 3 days after injury, and then tended to be flat. (U–X) The figures demonstrated the top 10 of pathway enrichment in blue, brown, green, and turquoise. 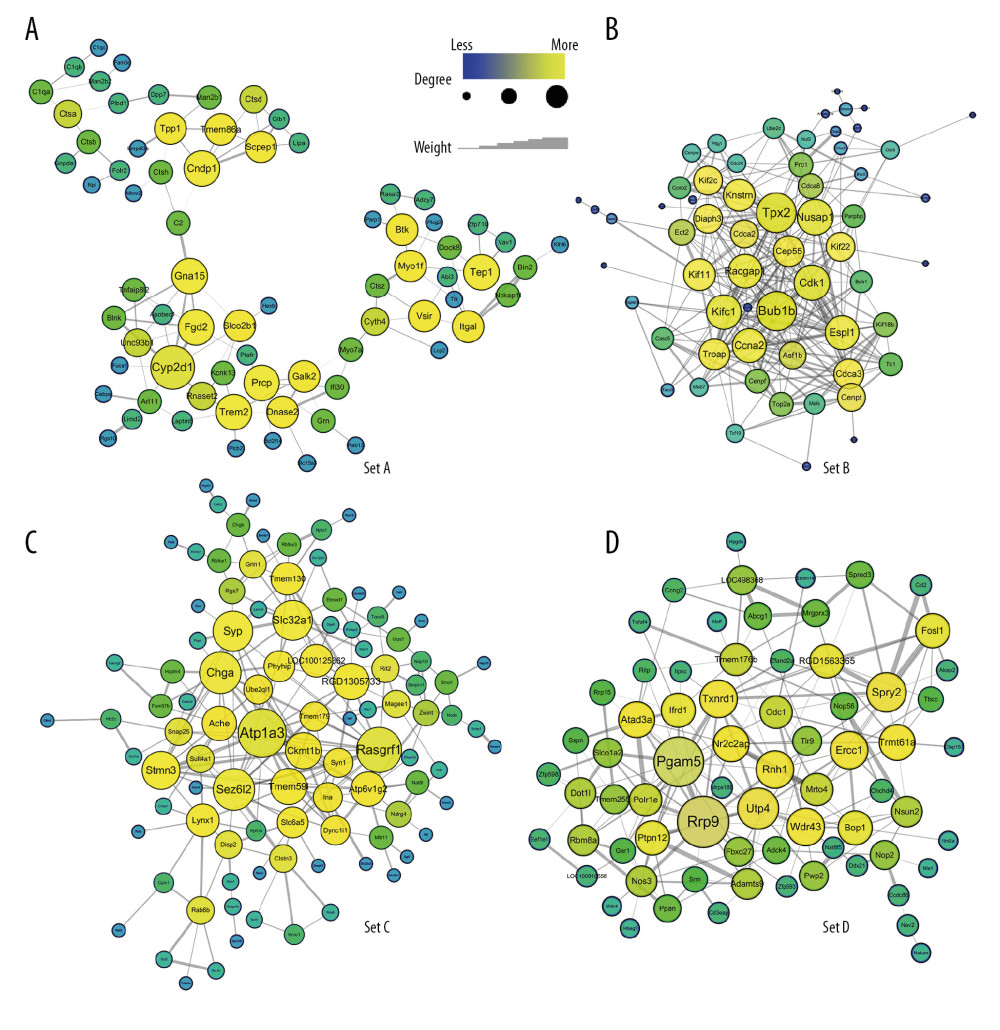 Figure 4. (A–D) Hub genes were identified in 4 sets through weighted gene coexpression network analysis (WGCNA). It was found that set A had 3 subparts, Cyp2d1 was the hub gene of set A, which was the core of the network, and C2 and Cyth4 were the key nodes that linked the other 2 parts. The other 3 sets exhibited their respective aggregations. Bub1b and Tpx2 were hub genes of set B. Similarly, Atp1a3 was the hub gene of set C, while Rrp9 and Pgam5 were hub genes of set D. Line thickness represented connection weight. The size and color of the nodes represented the degree of connectivity.
Figure 4. (A–D) Hub genes were identified in 4 sets through weighted gene coexpression network analysis (WGCNA). It was found that set A had 3 subparts, Cyp2d1 was the hub gene of set A, which was the core of the network, and C2 and Cyth4 were the key nodes that linked the other 2 parts. The other 3 sets exhibited their respective aggregations. Bub1b and Tpx2 were hub genes of set B. Similarly, Atp1a3 was the hub gene of set C, while Rrp9 and Pgam5 were hub genes of set D. Line thickness represented connection weight. The size and color of the nodes represented the degree of connectivity. References
1. Kumar R, Lim J, Mekary RA, Traumatic spinal injury: Global epidemiology and worldwide volume: World Neurosurg, 2018; 113; S1375005124
2. Alizadeh A, Dyck SM, Karimi-Abdolrezaee S, Traumatic spinal cord injury: An overview of pathophysiology, models and acute injury mechanisms: Front Neurol, 2019; 10; 282
3. Venkatesh K, Ghosh SK, Mullick M, Spinal cord injury: Pathophysiology, treatment strategies, associated challenges, and future implications: Cell Tissue Res, 2019; 377(2); 125-51
4. Zhang B, Horvath S, A general framework for weighted gene co-expression network analysis: Stat Appl Genet Mol Biol, 2005; 4; e17
5. Li A, Horvath S, Network neighborhood analysis with the multi-node topological overlap measure: Bioinformatics, 2007; 23(2); 222-31
6. Langfelder P, Horvath S, WGCNA: An R package for weighted correlation network analysis: BMC Bioinformatics, 2008; 9; 559
7. Li Z, Liu F, Zhang L, Neuroserpin restores autophagy and promotes functional recovery after acute spinal cord injury in rats: Mol Med Rep, 2018; 17(2); 2957-63
8. Chen Z, Li Z, Jiang C, MiR-92b-3p promotes neurite growth and functional recovery via the PTEN/AKT pathway in acute spinal cord injury: J Cell Physiol, 2019; 234(12); 23043-52
9. Ritchie ME, Phipson B, Wu D, limma powers differential expression analyses for RNA-sequencing and microarray studies: Nucleic Acids Res, 2015; 43(7); e47
10. Benjamini Y, Hochberg Y, Controlling the false discovery rate: A practical and powerful approach to multiple testing: J R Stat Series B Methodol, 1995; 57(1); 289-300
11. Shannon P, Markiel A, Ozier O, Cytoscape: A software environment for integrated models of biomolecular interaction networks: Genome Res, 2003; 13(11); 2498-504
12. The Gene Ontology Consortium, The Gene Ontology Resource: 20 years and still GOing strong: Nucleic Acids Res, 2019; 47(D1); D330-38
13. Ashburner M, Ball CA, Blake JA, Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium: Nat Genet, 2000; 25(1); 25-29
14. Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources: Nat Protoc, 2009; 4(1); 44-57
15. Huang DW, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists: Nucleic Acids Res, 2009; 37(1); 1-13
16. Dokalis N, Prinz M, Resolution of neuroinflammation: Mechanisms and potential therapeutic option: Semin Immunopathol, 2019; 41(6); 699-709
17. Wang Y, Bi Y, Xia Z, Butylphthalide ameliorates experimental autoimmune encephalomyelitis by suppressing PGAM5-induced necroptosis and inflammation in microglia: Biochem Biophys Res Commun, 2018; 497(1); 80-86
18. Moriwaki K, Farias LN, Balaji S, The mitochondrial phosphatase PGAM5 is dispensable for necroptosis but promotes inflammasome activation in macrophages: J Immunol, 2016; 196(1); 407-15
19. Liu S, Li Y, Choi H, Lysosomal damage after spinal cord injury causes accumulation of RIPK1 and RIPK3 proteins and potentiation of necroptosis: Cell Death Dis, 2018; 9(5); 476
20. Zhou X, Wahane S, Friedl M, Microglia and macrophages promote corralling, wound compaction and recovery after spinal cord injury via Plexin-B2: Nat Neurosci, 2020; 23(3); 337-50
21. Tyndale RF, Li Y, Li NY, Characterization of cytochrome P-450 2D1 activity in rat brain: High-affinity kinetics for dextromethorphan: Drug Metab Dispos, 1999; 27(8); 924-30
22. Ma J, Wang J, Cheng J, Impacts of blast-induced traumatic brain injury on expressions of hepatic cytochrome P450 1A2, 2B1, 2D1, and 3A2 in Rats: Cell Mol Neurobiol, 2017; 37(1); 111-20
23. Tica J, Bradbury EJ, Didangelos A, Combined transcriptomics, proteomics and bioinformatics identify drug targets in spinal cord injury: Int J Mol Sci, 2018; 19(5); 1461
24. Carecchio M, Zorzi G, Ragona F, ATP1A3-related disorders: An update: Eur J Paediatr Neurol, 2018; 22(2); 257-63
25. Chu Y, Parada I, Prince DA, Temporal and topographic alterations in expression of the alpha3 isoform of Na+, K(+)-ATPase in the rat freeze lesion model of microgyria and epileptogenesis: Neuroscience, 2009; 162(2); 339-48
26. Patricia DCA, Sweadner KJ, Penniston JT, Mutations in the Na+/K+ -ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism: Neuron, 2004; 43(2); 169-75
27. Silver J, Miller JH, Regeneration beyond the glial scar: Nature Rev Neurosci, 2004; 5(2); 146-56
28. Bradbury EJ, Burnside ER, Moving beyond the glial scar for spinal cord repair: Nature Commun, 2019; 10(1); 3879
29. Izumi H, Matsumoto Y, Ikeuchi T, BubR1 localizes to centrosomes and suppresses centrosome amplification via regulating Plk1 activity in interphase cells: Oncogene, 2009; 28(31); 2806-20
30. Del Bene F, Interkinetic nuclear migration: Cell cycle on the move: EMBO J, 2011; 30(9); 1676-77
31. Scrofani J, Sardon T, Meunier S, Microtubule nucleation in mitosis by a RanGTP-dependent protein complex: Curr Biol, 2015; 25(2); 131-40
32. He J, Wu J, Xu N, MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes: Nucleic Acids Res, 2013; 41(1); 498-508
33. Zissler A, Steinbacher P, Zimmermann R, Extracorporeal shock wave therapy accelerates regeneration after acute skeletal muscle injury: Am J Sports Med, 2017; 45(3); 676-84
34. Bazrafshan A, Owji M, Yazdani M, Activation of mitosis and angiogenesis in diabetes-impaired wound healing by processed human amniotic fluid: J Surg Res, 2014; 188(2); 545-52
35. Wu J, Stoica BA, Faden AI, Cell cycle activation and spinal cord injury: Neurotherapeutics, 2011; 8(2); 221-28
36. Di Giovanni S, Knoblach SM, Brandoli C, Gene profiling in spinal cord injury shows role of cell cycle in neuronal death: Ann Neurol, 2003; 53(4); 454-68
37. Wang T, Wu B, Zhang X, Identification of gene coexpression modules, hub genes, and pathways related to spinal cord injury using integrated bioinformatics methods: J Cell Biochem, 2019; 120(5); 6988-97
38. Duan H, Ge W, Zhang A, Transcriptome analyses reveal molecular mechanisms underlying functional recovery after spinal cord injury: Proc Natl Acad Sci USA, 2015; 112(43); 13360-65
Figures
 Figure 1. Establishment of spinal cord injury after compression. (A) Diagram of compression model of spinal cord injury (SCI). (B–G) The figure demonstrated the visible signs of bleeding and spinal cord disruption after SCI. (H–Q) H&E staining showed the pathological changes at different time points (Sham, 1d, 3d, 5d, and 7d) after SCI in, coronal, and transverse sections (white arrows mark the injury site).
Figure 1. Establishment of spinal cord injury after compression. (A) Diagram of compression model of spinal cord injury (SCI). (B–G) The figure demonstrated the visible signs of bleeding and spinal cord disruption after SCI. (H–Q) H&E staining showed the pathological changes at different time points (Sham, 1d, 3d, 5d, and 7d) after SCI in, coronal, and transverse sections (white arrows mark the injury site). Figure 2. Differential gene screening and weighted gene coexpression network analysis. (A) A total of 1965 differential genes were screened at different time points (Sham, 1d, 3d, 5d, and 7d) after spinal cord injury (SCI). (B, C) Analysis of network topology showed that it met the scale-free topology threshold of 0.8 when β=16. (D) The heatmap of topological matrix showed the correlation network, in which each color represented a gene module.
Figure 2. Differential gene screening and weighted gene coexpression network analysis. (A) A total of 1965 differential genes were screened at different time points (Sham, 1d, 3d, 5d, and 7d) after spinal cord injury (SCI). (B, C) Analysis of network topology showed that it met the scale-free topology threshold of 0.8 when β=16. (D) The heatmap of topological matrix showed the correlation network, in which each color represented a gene module. Figure 3. Expression trends and pathway enrichment of modules identified via WGCNA. (A–T) A total of 10 modules were split and each module was assigned a symbolic color. It was found that the modules of magenta and brown were sharply downregulated on the first day after injury, then tended to recover; the modules of blue, pink, green, grey, and red were downregulated after injury, and then rebounded upward 3 days after SCI; the modules of black and turquoise showed a continuous decrease after injury; the yellow module was sharply upregulated within 3 days after injury, and then tended to be flat. (U–X) The figures demonstrated the top 10 of pathway enrichment in blue, brown, green, and turquoise.
Figure 3. Expression trends and pathway enrichment of modules identified via WGCNA. (A–T) A total of 10 modules were split and each module was assigned a symbolic color. It was found that the modules of magenta and brown were sharply downregulated on the first day after injury, then tended to recover; the modules of blue, pink, green, grey, and red were downregulated after injury, and then rebounded upward 3 days after SCI; the modules of black and turquoise showed a continuous decrease after injury; the yellow module was sharply upregulated within 3 days after injury, and then tended to be flat. (U–X) The figures demonstrated the top 10 of pathway enrichment in blue, brown, green, and turquoise. Figure 4. (A–D) Hub genes were identified in 4 sets through weighted gene coexpression network analysis (WGCNA). It was found that set A had 3 subparts, Cyp2d1 was the hub gene of set A, which was the core of the network, and C2 and Cyth4 were the key nodes that linked the other 2 parts. The other 3 sets exhibited their respective aggregations. Bub1b and Tpx2 were hub genes of set B. Similarly, Atp1a3 was the hub gene of set C, while Rrp9 and Pgam5 were hub genes of set D. Line thickness represented connection weight. The size and color of the nodes represented the degree of connectivity.
Figure 4. (A–D) Hub genes were identified in 4 sets through weighted gene coexpression network analysis (WGCNA). It was found that set A had 3 subparts, Cyp2d1 was the hub gene of set A, which was the core of the network, and C2 and Cyth4 were the key nodes that linked the other 2 parts. The other 3 sets exhibited their respective aggregations. Bub1b and Tpx2 were hub genes of set B. Similarly, Atp1a3 was the hub gene of set C, while Rrp9 and Pgam5 were hub genes of set D. Line thickness represented connection weight. The size and color of the nodes represented the degree of connectivity. Tables
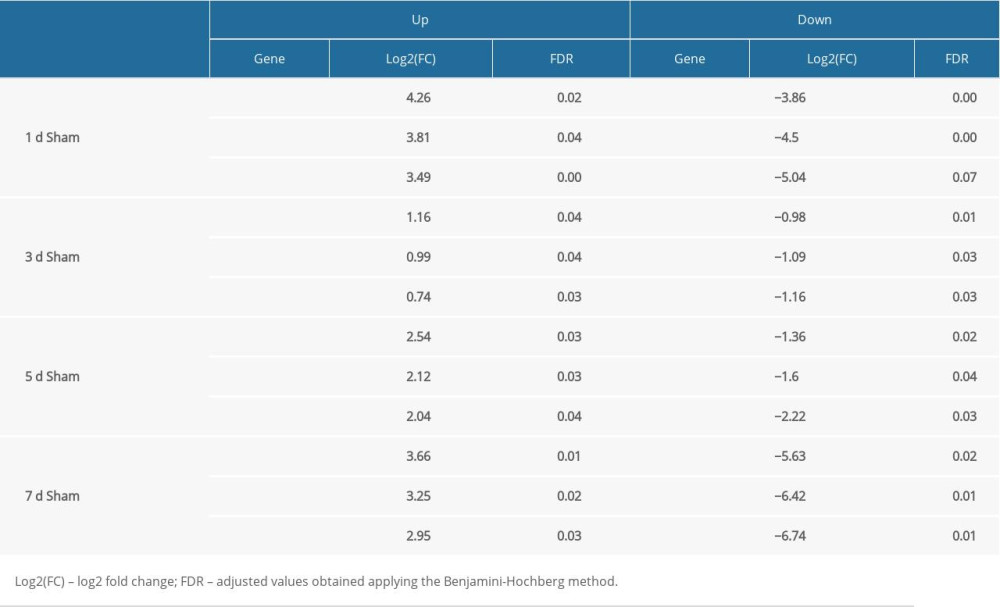 Table 1. Top 3 up or down genes by comparing average FPKM between different days post-injury and Sham group.
Table 1. Top 3 up or down genes by comparing average FPKM between different days post-injury and Sham group.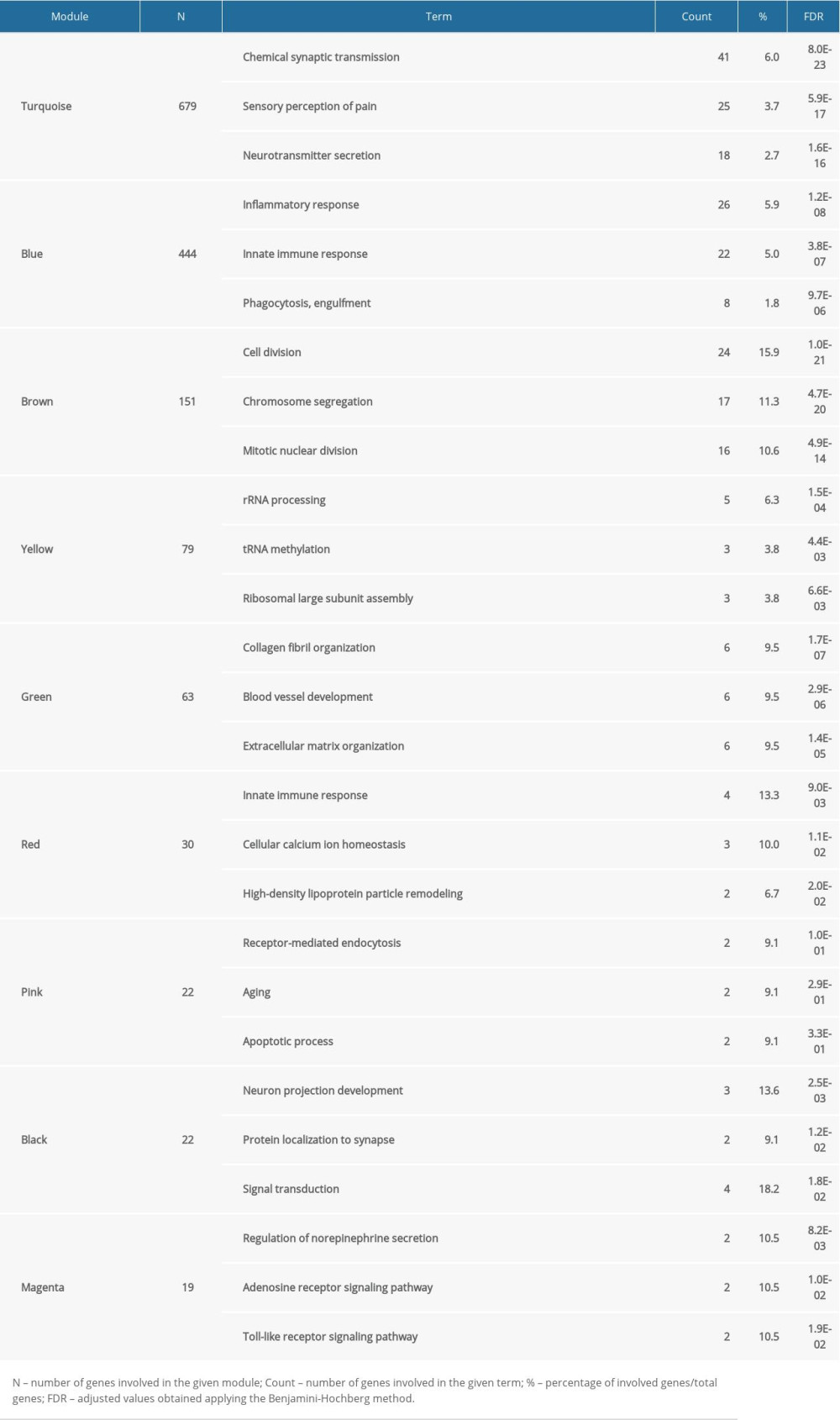 Table 2. Overview of 10 modules with their top 3 GO-BP terms.
Table 2. Overview of 10 modules with their top 3 GO-BP terms.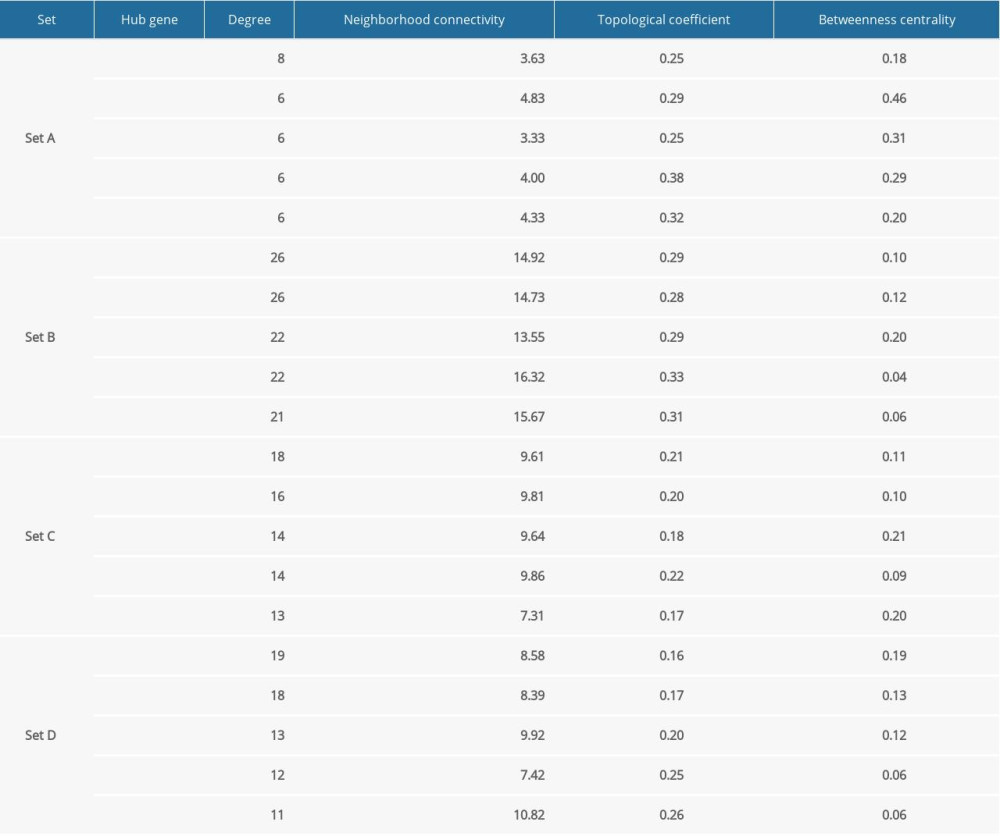 Table 3. Hub genes of module sets.
Table 3. Hub genes of module sets. Table 1. Top 3 up or down genes by comparing average FPKM between different days post-injury and Sham group.
Table 1. Top 3 up or down genes by comparing average FPKM between different days post-injury and Sham group. Table 2. Overview of 10 modules with their top 3 GO-BP terms.
Table 2. Overview of 10 modules with their top 3 GO-BP terms. Table 3. Hub genes of module sets.
Table 3. Hub genes of module sets. In Press
05 Mar 2024 : Clinical Research
Effects of Thermal Insulation on Recovery and Comfort of Patients Undergoing Holmium Laser LithotripsyMed Sci Monit In Press; DOI: 10.12659/MSM.942836
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








