21 October 2021: Clinical Research
Integrating Intrinsic Radiosensitivity and Immune Status for Predicting Benefits of Radiotherapy in Head and Neck Squamous Cell Carcinoma
Chuyao Sun12ADE, Miao Zhang1BCF, Qiao Qiao1ADE, Yanli Wang1C*DOI: 10.12659/MSM.932126
Med Sci Monit 2021; 27:e932126
Abstract
BACKGROUND: HNSCC (head and neck squamous cell carcinoma) is a heterogeneous disease for which radiotherapy is a main treatment. As intrinsic radiosensitivity and immune status affect the initial and effective stage of the radiation-induced cancer immunity cycle, respectively, it is important to consider both of them when we select patients who can benefit from radiotherapy.
MATERIAL AND METHODS: Our study included all HNSCC patients with complete survival and radiotherapy information in TCGA database. Patients were divided into RS (radiosensitive), RR (radioresistant), immune, and non-immune groups according to their RSI (radiosensitivity index) and immune score calculated by the ESTIMATE algorithm. Survival analysis was performed to compare OS (overall survival) between patients receiving and not receiving radiotherapy. GO and KEGG pathway enrichment analysis was performed for functional analysis. Univariate Cox and ridge regression analysis were performed to construct a predictive gene signature based on the combined stratification.
RESULTS: Only patients in the RS-immune group could benefit from radiotherapy, and the survival analysis results remained consistent after we performed propensity score matching between patients receiving and not receiving radiotherapy. The differentially expressed genes between the RS-immune and non-RS-immune groups were mainly enriched in pathways related to immune process. The 3-gene signature we built exhibited predictive value in training and validation cohorts when treated as a binary or continuous variable.
CONCLUSIONS: The combined stratification of intrinsic radiosensitivity and immune status was superior to considering intrinsic radiosensitivity or immune status alone and could be used in preclinical evaluation to select patients or to decide whether radiotherapy sensitizers and immunotherapy should be used at the same time.
Keywords: Immunity, Active, Radiotherapy, Squamous Cell Carcinoma of Head and Neck, Biomarkers, Tumor, Head and Neck Neoplasms, Humans, Immunotherapy, Radiation Tolerance
Background
HNSCC (head and neck squamous cell carcinoma) is the sixth most common cancer in the world [1]. The incidence of HNSCC varies from region to region, and is generally associated with the exposure to tobacco-derived carcinogens or excessive alcohol consumption [2]. Over 600 000 new cases and 350 000 deaths occurred in a year. The 5-year survival rate of patients with locally advanced HNSCC is about 50% and 40–60% of patients relapse and do not respond to subsequent treatment. The median survival time of patients with recurrent/metastatic disease is only 10 months [3,4]. Radiotherapy is one of the main treatments for HNSCC; however, because of the complexity and heterogeneity of HNSCC, not all patients will benefit from radiotherapy. Some patients cannot tolerate the adverse reactions, resulting in treatment termination or even death [5–8]. Therefore, biomarkers for predicting radiosensitivity in HNSCC are necessary for radiotherapy selection and predicting therapeutic response.
In recent years, some studies have proposed use of gene expression signatures to predict survival in HNSCC patients receiving radiotherapy. For example, Eschrich et al [9] developed the intrinsic radiosensitivity index (RSI) through the SF2 of 48 cancer cell lines, and RSI has already been systematically validated as a predictor of clinical outcomes in multiple independent radiotherapy cohorts including head and neck cancer [10]. Ma et al identified a 4-gene methylation signature to predict survival in HNSCC patients [11]. You et al identified 4 key markers in 4 core functional pathways representing radiosensitivity of HNSCC [12]. However, although most of the existing signatures are prognostically relevant, few have been shown to be predictive in HNSCC.
Previous studies on improving radiotherapy effects mostly focused only on cancer cells, but there is now growing evidence that the therapeutic effect of radiotherapy is influenced and regulated by the tumor microenvironment, including the immune system [13]. In the radiation-induced cancer immunity cycle, intrinsic radiosensitivity affects the release of cancer cell antigen, and immune status affects antigen-specific T cell activation [14]. In this work, we explored the effect of combined stratification of intrinsic radiosensitivity and immune status on the benefit of radiotherapy in HNSCC patients by integrating 2 different biological processes. Based on this combined stratification, a predictive gene expression signature was established to select patients who could benefit from radiotherapy.
Material and Methods
DATA ACQUISITION AND PROCESSING:
HNSCC clinical and raw gene count expression data were downloaded from TCGA (http://cancergenome.nih.gov/, 2020/03/15). Patients with both survival and radiotherapy information were retained (n=392). Genes with maximum expression values less than 10 and average expression values less than 1 were excluded because their expression values were too low. The DEseq2 package [15] was used to standardize raw gene expression data and change gene expression to log2 (count+1) form. All protein coding genes were extracted for subsequent analysis. Immune scores calculated by ESTIMATE algorithm was downloaded from the website (https://bioinformatics.mdanderson.org/estimate/, 2020/03/31).
PATIENT STRATIFICATION AND SURVIVAL ANALYSIS:
Patients were divided into an immune group and a non-immune group based on the median immune score. RSI value of each patient was calculated based on the gene model established by Eschrich et al [9]. We selected patients who received radiotherapy (N=254) and determined the optimal cut-off value as the RSI value that could separate those patients into 2 groups with the most significant difference in OS using X-tile software (version 3.6.1, Yale University School of Medicine, New Haven, CT, USA). All patients were divided into an RS group (N=157), an RR group (N=235), an immune group (N=196), and a non-immune (N=196) group. We compared overall survival between patients receiving and not receiving radiotherapy in the 4 groups. After that, patients were divided into 4 subgroups: RS-immune (N=83), RS-non-immune (N=74), RR-immune (N=113), and RR-non-immune (N=122). The overall survival of patients receiving radiotherapy and not receiving radiotherapy was compared in the 4 subgroups.
PATIENT MATCHING AND SURVIVAL ANALYSIS:
To minimize selection bias and confounding factors, we performed propensity score matching (PSM) between patients receiving and not receiving radiotherapy. We considered RS-immune as 1 group and the 3 other groups as another group, called the non-RS-immune group. Because of the lack of clinical information, we matched patients with a ratio of 1: 4 based on 7 clinical and therapeutic features including neoplasm histological grade (grade I, II, III), clinical stage (stage I, II, III, IV), lymphovascular invasion (no, yes), margin status (negative, positive\approach), presence of pathological nodal extra-capsular spread (no, micro-invasion\ gross invasion), surgery (no, yes), and chemotherapy (no, yes). After that, overall survival of patients receiving radiotherapy and not receiving radiotherapy was compared in 2 matching cohorts.
GENE DIFFERENTIAL EXPRESSION ANALYSIS AND PATHWAY ENRICHMENT ANALYSIS:
DEG (differentially expressed genes) between RS-immune and non-RS-immune group were identified with a cut-off value of adjust p value <0.05 and |log2 fold change (FC)| ≥1 using Limma package [16]. GO (Gene Ontology) biological process and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis was performed using DAVID website (https://david.ncifcrf.gov/) with a selection criterion of FDR<0.05.
GENE SIGNATURE ESTABLISHMENT WITH RIDGE-REGULARIZED COX MODEL:
To establish a gene expression model that can predict the benefit of radiotherapy, we randomly divided the TCGA dataset into a training (N=236) and a validation cohort (N=156) with a ratio of 3: 2. We analyzed the DEG between the RS-immune and non-RS-immune group. Genes passed the selection criteria of |log2FC| ≥1.2, and adjusted P value <0.01 were selected for subsequent survival analysis. We use the method developed by Tian et al [13,17] to train this predictive gene model with the Penalized package [18]. First, we performed univariate Cox regression analyses with the formula below:
X represents the expression of DEGs. T represents radiotherapy status and was coded as {−1,1}. Genes having significant interaction with radiotherapy (
X1….Xk represents the genes selected by univariate Cox regression analyses and lambda represents the regularization intensity determined by 10-fold cross-validation. The final gene signature for RSS was:
STATISTICAL ANALYSIS:
X-tile software (version 3.6.1) was used to calculate RSI best cut-off value, and all other statistical analysis were performed by R Studio software (version 1.1.453). Kaplan-Meier survival analysis and Cox regression analyses were used for survival analysis. The chi-square test and Fisher exact test were used to compare clinical and pathological factors between corresponding groups. All tests were 2-sided. A
Results
PATIENT STRATIFICATION AND SURVIVAL ANALYSIS:
The RSI cut-off value performed by X-tile software was 0.8. Schema of patient grouping is provided in Figure 1. Based on the cut-off value of RSI and the median value of immune score, we divided the cohort into the RS group and RR group, and the immune group and non-immune group. Kaplan-Meier survival analysis suggested that better overall survival in patients receiving radiotherapy were seen in the RS and immune groups (Figure 2A, P<0.001, HR=0.338, 95% CI=0.187–0.611; Figure 2C, P=0.003, HR=0.482, 95% CI=0.296–0.785). On the contrary, there were no significant difference in overall survival between patients receiving and not receiving radiotherapy in RR or in the non-immune group (Figure 2B, P=0.313, HR=0.814, 95% CI=0.545–1.214; Figure 2D, P=0.197, HR=0.742, 95% CI=0.472–1.167)
After that, we combined the 2 stratification methods to divide patients into 4 subgroups – the RS-immune, RS-non-immune, RR-immune, and RR-non-immune groups – showing that patients treated with radiotherapy had better overall survival compared with those without radiotherapy only in the RS-immune group (Figure 3A, P<0.001, HR=0.194, 95% CI=0.788–0.480). Among the rest groups, there were no significant differences in overall survival between patients receiving radiotherapy and not receiving radiotherapy (Figure 3B, P=0.276, HR=0.595, 95% CI=0.234–1.515; Figure 3C, P=0.571, HR=0.836, 95% CI=0.450–1.555; Figure 3D, P=0.492, HR=0.831, 95% CI=0.490–1.409).
PATIENTS MATCHING AND SURVIVAL ANALYSIS:
In order to exclude the influence of confounding factors, we performed PSM between patients receiving and not receiving radiotherapy. After PSM, RS-immune group remained 23 patients, non-RS-immune group remained 105 patients. There were no significant differences in the 7 matching variables between patients treated with and without radiotherapy in both groups (Tables 1, 2).
We performed survival analysis in the matching cohorts and found similar results to that before PSM. In the RS-immune group, the overall survival was better in patients receiving radiotherapy (Figure 4A, P=0.015, HR=0.102, 95% CI=0.011–0.933). In the non-RS-immune group, there was no significant difference in overall survival between patients receiving and not receiving radiotherapy (Figure 4B, P=0.112, HR=0.600, 95% CI=0.320–1.126).
BIOINFORMATION ANALYSIS OF DIFFERENTIALLY EXPRESSED GENES:
To explore potential biological processes, we analyzed the differentially expressed genes between RS-immune group and non-RS-immune group. The results showed that there were 674 differentially expressed genes with 525 genes upregulated in RS-immune group and 149 downregulated. GO and KEGG pathway enrichment analysis were performed using differentially expressed genes. Top 10 GO biological processes and KEGG pathways were shown (Figure 5A, 5B). GO biological process enrichment analysis showed that the differentially expressed genes were mainly enriched in pathways related to immunity such as immune response, adaptive immune response, and T cell co-stimulation, suggesting an active immune response in the RS-immune group. KEGG pathway enrichment results showed that the differentially expressed genes were mainly enriched in cytokine-cytokine receptor interaction, intestinal immune network for IgA production, and cell adhesion molecules.
PREDICTIVE VALUE OF THE SIGNATURE IN HNSCC:
To establish a gene expression model that can predict radiotherapy benefit, we divided the TCGA dataset into a training and a validation cohort. The information of the 2 cohorts is shown in Table 3. The differentially expressed genes analysis in the training cohort identified 382 genes, all of which were included in the univariate Cox regression analyses. The results showed that 3 genes had significant interactions with radiotherapy (P<0.05) and they were further included in the multivariate ridge-regularized Cox model. We successfully constructed a 3-gene signature: RSS=0.0352306203* CCNA1-0.0366707616* CLGN-0.0224280179* KRT15, lambda=674.5671(Figure 6).
In the training cohort, we used the median value of RSS to divide the cohort into 2groups; the RSS-L group was classified as the radiotherapy-effective group because patients receiving radiotherapy had better OS (Figure 7A, P=0.001, HR=0.377, 95% CI=0.202–0.703), while patients in the RSS-H group were classified as radiotherapy-defective as no significant survival difference could be seen between patients receiving and not receiving radiotherapy (Figure 7B, P=0.634, HR=1.156, 95% CI=0.636–2.103). The validation cohort was also split into 2 groups according to RSS median value. Consistent with the result of the training set, patients in RSS-L group benefited from radiotherapy (Figure 7C, P<0.001, HR=5.215, 95% CI=2.235–12.166) and there was no significant difference in overall survival between patients treated with or without radiotherapy in the RSS-H group (Figure 7D, P=0.915, HR=1.041, 95% CI=0.494–2.196). Finally, we considered RSS as a continuous variable and performed univariate and multivariate Cox regression analyses in the training and validation cohorts. RSS showed significant interaction with radiotherapy in univariate and multivariate analyses in the training and validation cohorts (Table 4).
Discussion
In this study, we first demonstrated that the combined stratification of intrinsic radiosensitivity and immune status could predict radiotherapy benefit and was better than considering intrinsic radiosensitivity or immune status alone. In addition, we developed a gene signature based on the combined stratification for predicting radiotherapy benefit of HNSCC independently.
Given the heterogeneity of HNSCC, we need effective biomarkers to select dominant patients and guide the optimal use of radiotherapy. At present, clinical and pathological parameters such as TNM stage, lymph node extra-capsular invasion, lymph vascular invasion, surgical margin, and nerve invasion are the main factors affecting radiotherapy decision-making in HNSCC. However, none of these parameters fully explain the differences in treatment response for individual patients undergoing radiotherapy. Other patient characteristics, such as smoking status, age, and nutritional status [19,20] are also considered to be factors that influence radiotherapy response and prognosis, but it is still difficult to predict which patients will be completely relieved and which will not. This condition makes the treatment of a large number of patients inadequate, ineffective, or unnecessary.
In this paper, we first demonstrate that the intrinsic radiosensitivity affects radiation response, which is widely accepted. Then, we demonstrate that patients with high immune score are more likely to benefit from radiotherapy. Previous articles have proved that immune cells in the tumor microenvironment are necessary for sufficient radiation response in preclinical mouse models [21–23]. Kawaguchi et al [24] put forward that high CD8+ TIL density is a favorable biomarker for patients with nasopharyngeal carcinoma or oropharyngeal carcinoma. Alternatively, Gurin et al [25] also suggested that patients with high density of stromal CD8+ TILs displayed significantly better overall survival (OS) and progression-free survival (PFS) in oropharyngeal squamous cell carcinoma patients receiving IMRT. However, there are limited articles combing these 2 factors together. Patel et al [26] demonstrated that the combination of radiation, G2/M cell cycle checkpoint regulator Wee1 kinase inhibitor ADZ1775 and immunomodulator PD-1 can induce rejection of established tumor in 73% of the oral cancer in a mouse model, which is better than radiation or ADZ1775 dual therapy with PD-1 and is also superior to PD-1 monoclonal antibody monotherapy. This is consistent with our results. Our results indicate that only patients who are intrinsically radiosensitive and have abundant immune cells infiltration can benefit from radiotherapy. The possible explanation is that the radiation-induced cancer immunity cycle kills tumor cells by inducing tumor cell death, triggering innate immune system, activating tumor specific T cells that can bind to tumor cells, killing cancer cells, releasing other tumor related antigens, and repeating and expanding the above process again [14]. Tumor antigen releasing is the first step to initiate anti-cancer immunity, and radiotherapy can accelerate this step by inducing immunogenic cell death [27]. Patients who are not sensitive to radiotherapy have weaker response; therefore, less tumor antigens are released and this may result in insufficient CTL infiltration. On the other hand, the presence of antigen-presenting defects, depletion of T cells, tumor insensitivity to T cells, immunosuppressive cells, and other immunosuppressive checkpoints in the tumor microenvironment may also weaken cytotoxic effect of CTL [14,28]. The complementary effect of the 2 biological processes makes intrinsic radiosensitivity and immune status become 2 essential conditions for patients to benefit from radiotherapy. Through our conclusion, we can identify patients who may not respond well to standard radiotherapy and may benefit from radiation sensitizers or immunotherapy.
Some gene expression-based signatures have been constructed to predict the overall survival of HNSCC patients treated with RT. Ma et al [11] established a 4-gene methylation signature to predict survival rate of HNSCC patients with RT, and You et al [12] identified 4 key molecular markers based on 4 core functional pathways. These markers are prognostic at the time of establishment. However, we established a predictive model, RSS, by calculating the interaction between gene expression and radiotherapy with the method of Tian et al [17]. As a result, our signature may be more suitable to help make treatment decisions in clinical practice.
The main limitation of our study was the combination of retrospective cohort and non-randomized radiotherapy. Ideally, predictive biomarkers should be tested in prospective randomized controlled trials. To reduce the potential selection bias in retrospective cohort studies, we used PSM to balance the relevant clinical and pathology characteristics between the corresponding groups [13]. Another limitation is that the constructed gene signature needs further biological and clinical verification as biomarkers of radiosensitivity in HNSCC.
Conclusions
In conclusion, we proved that the combined stratification of intrinsic radiosensitivity and immune status could serve as a preclinical evaluation to identify patients benefiting from radiotherapy and helped patients to choose the best treatment strategy. Based on this combined stratification, a gene signature was established to independently predict radiotherapy benefits in HNSCC.
Figures
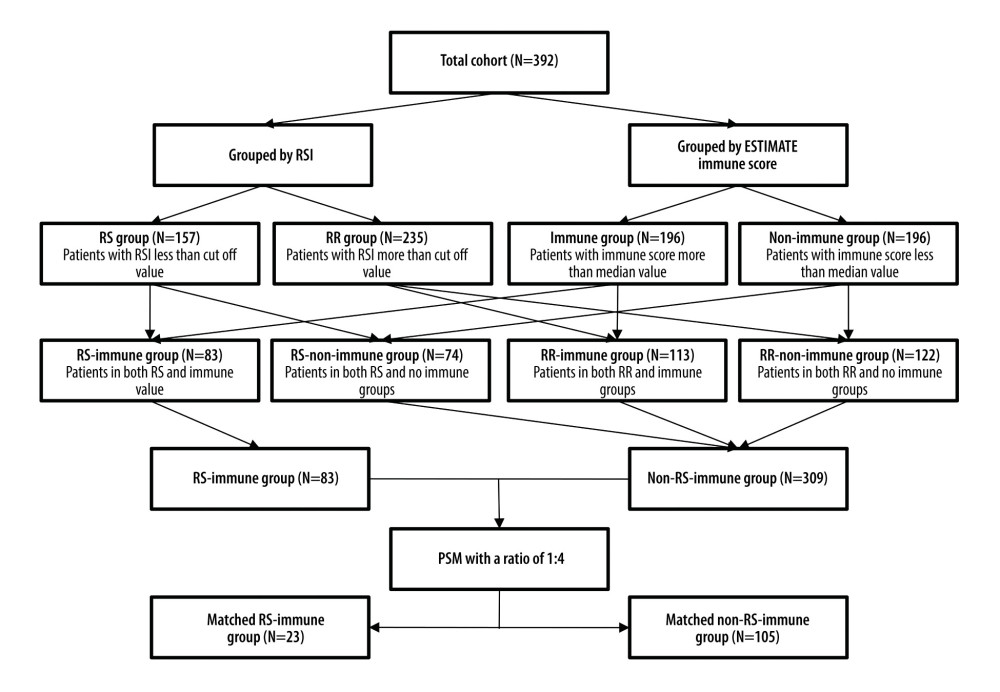 Figure 1. Schema of patient grouping. RSI – radiosensitivity index; RS – radiosensitive; RR – radioresistant; PSM – propensity score matching.
Figure 1. Schema of patient grouping. RSI – radiosensitivity index; RS – radiosensitive; RR – radioresistant; PSM – propensity score matching. 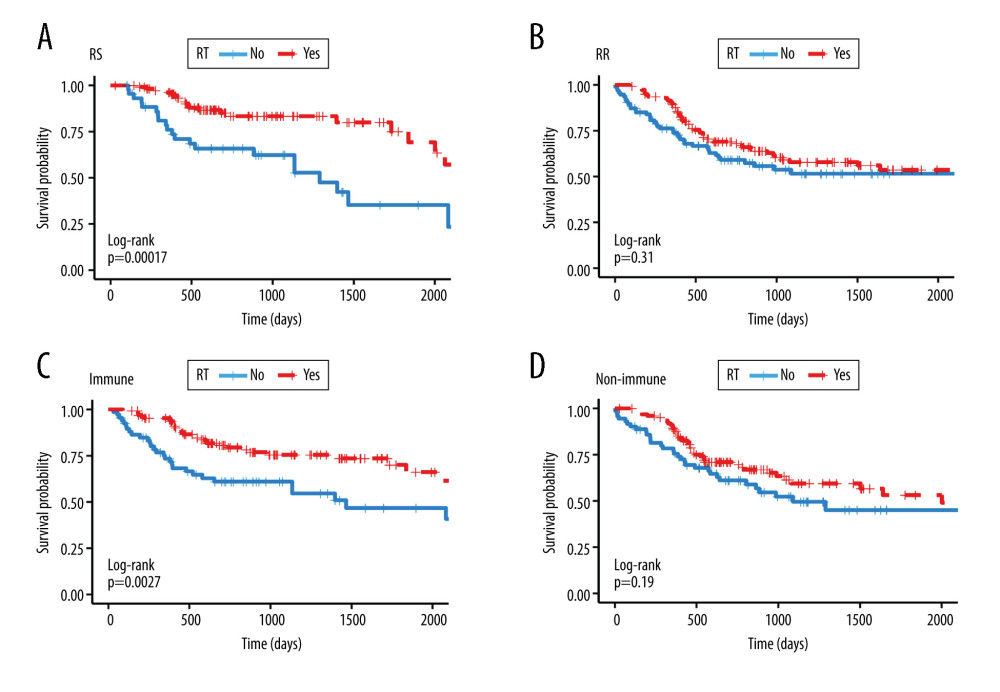 Figure 2. Kaplan-Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the group of (A) RS (B) RR (C) immune (D) non-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy.
Figure 2. Kaplan-Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the group of (A) RS (B) RR (C) immune (D) non-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy. 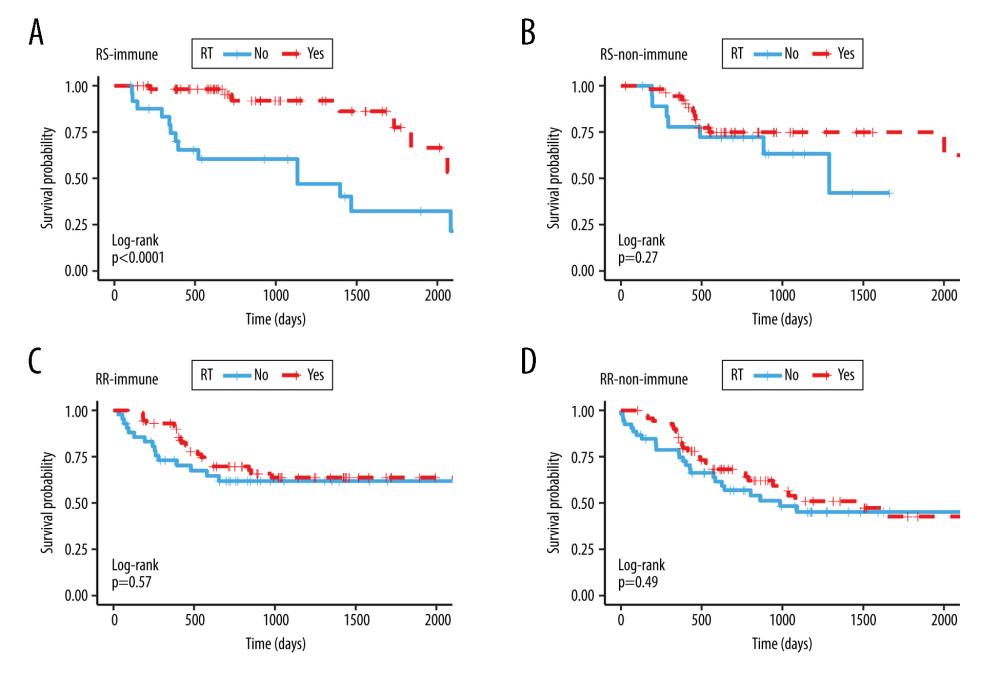 Figure 3. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the subgroup of (A) RS-immune (B) RS-non-immune (C) RR-immune (D) RR-non-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy.
Figure 3. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the subgroup of (A) RS-immune (B) RS-non-immune (C) RR-immune (D) RR-non-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy. 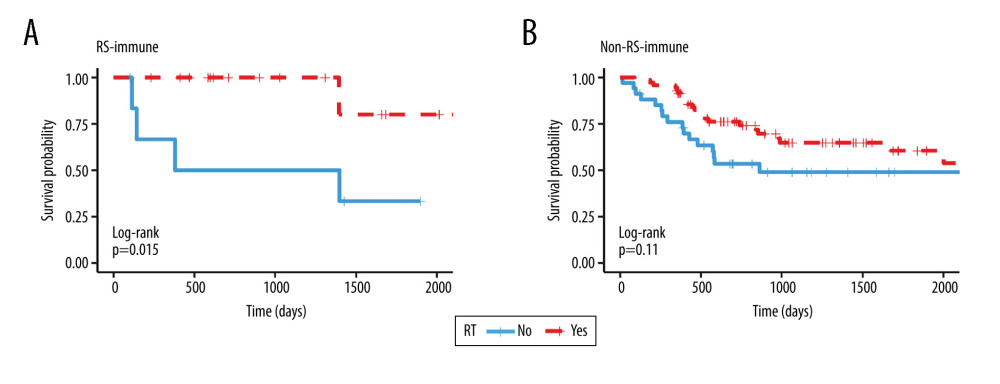 Figure 4. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the matching group of (A) RS-immune (B) non-RS-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy.
Figure 4. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the matching group of (A) RS-immune (B) non-RS-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy. 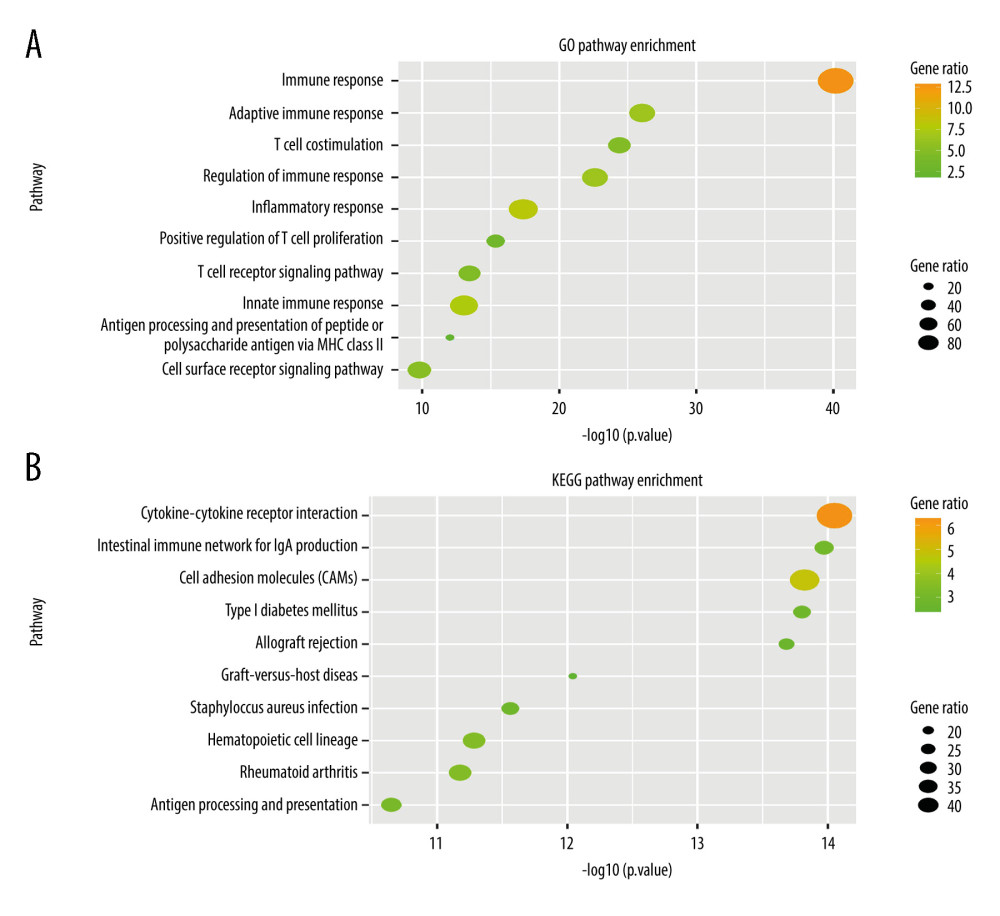 Figure 5. Gene Ontology and KEGG pathway enrichment analysis of differentially expressed genes between the RS-immune group and non-RS-immune group. The top 10 GO biological process enrichment results (A) and top 10 KEGG enrichment results (B) for differentially expressed genes.
Figure 5. Gene Ontology and KEGG pathway enrichment analysis of differentially expressed genes between the RS-immune group and non-RS-immune group. The top 10 GO biological process enrichment results (A) and top 10 KEGG enrichment results (B) for differentially expressed genes. 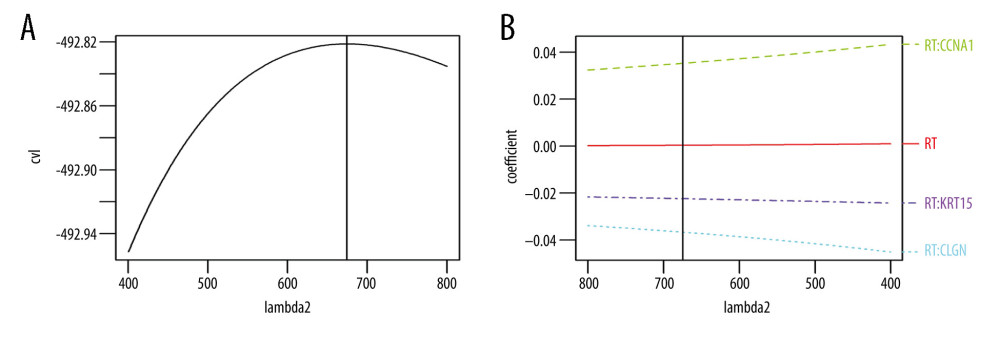 Figure 6. Signature established by L2 regularized (ridge) Cox regression model. (A) Parameter (Lambda2) selection by L2 regularized (ridge) Cox regression model adopted 10-fold cross-validation; (B) L2 regularized (ridge) Cox regression model coefficient profile plot of interaction between radiotherapy and genes selected by univariate Cox regression analyses. cvl – cross-validated likelihood; RT – radiotherapy.
Figure 6. Signature established by L2 regularized (ridge) Cox regression model. (A) Parameter (Lambda2) selection by L2 regularized (ridge) Cox regression model adopted 10-fold cross-validation; (B) L2 regularized (ridge) Cox regression model coefficient profile plot of interaction between radiotherapy and genes selected by univariate Cox regression analyses. cvl – cross-validated likelihood; RT – radiotherapy. 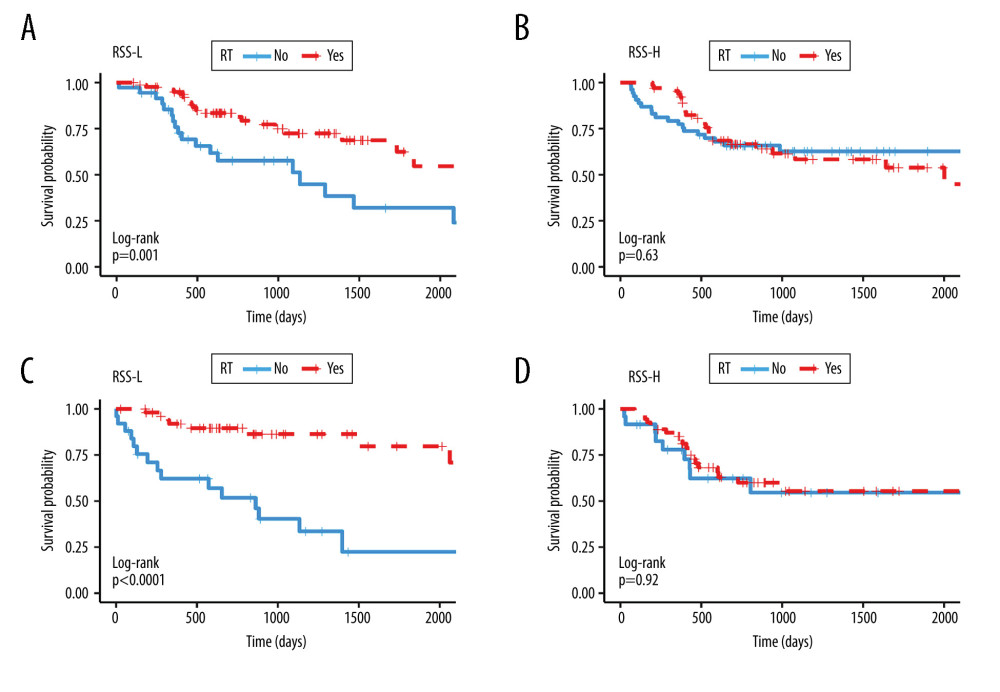 Figure 7. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the group of (A) RSS-L (B) RSS-H in the training cohort and in the group of (C) RSS-L (D) RSS-H in validation cohort. The median values of the RSS score were used as the cut offs to define the high and low RSS groups in both training and validation cohorts. RRS-L – RSS low expression; RSS-H – RSS high expression; RT – radiotherapy.
Figure 7. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the group of (A) RSS-L (B) RSS-H in the training cohort and in the group of (C) RSS-L (D) RSS-H in validation cohort. The median values of the RSS score were used as the cut offs to define the high and low RSS groups in both training and validation cohorts. RRS-L – RSS low expression; RSS-H – RSS high expression; RT – radiotherapy. Tables
Table 1. PSM result in RS-immune group.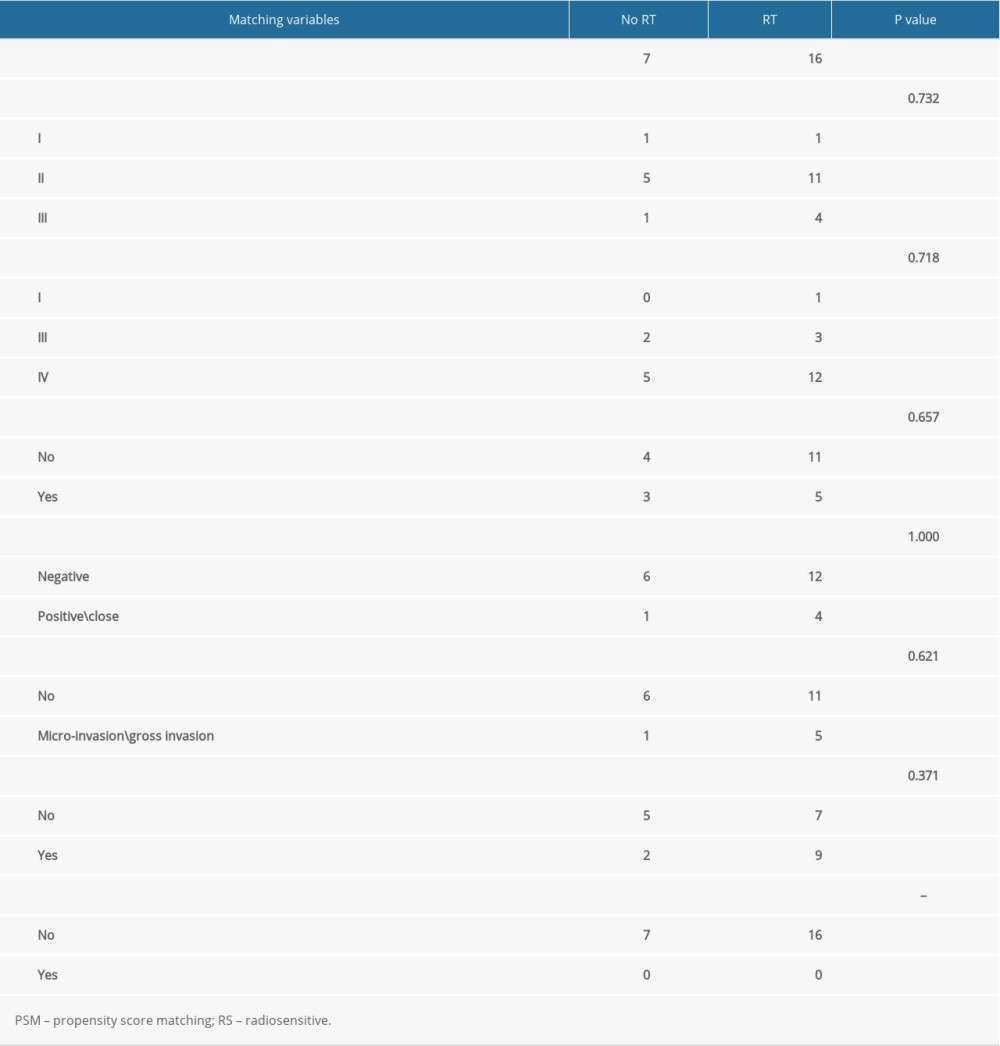 Table 2. PSM result in non-RS-immune group.
Table 2. PSM result in non-RS-immune group.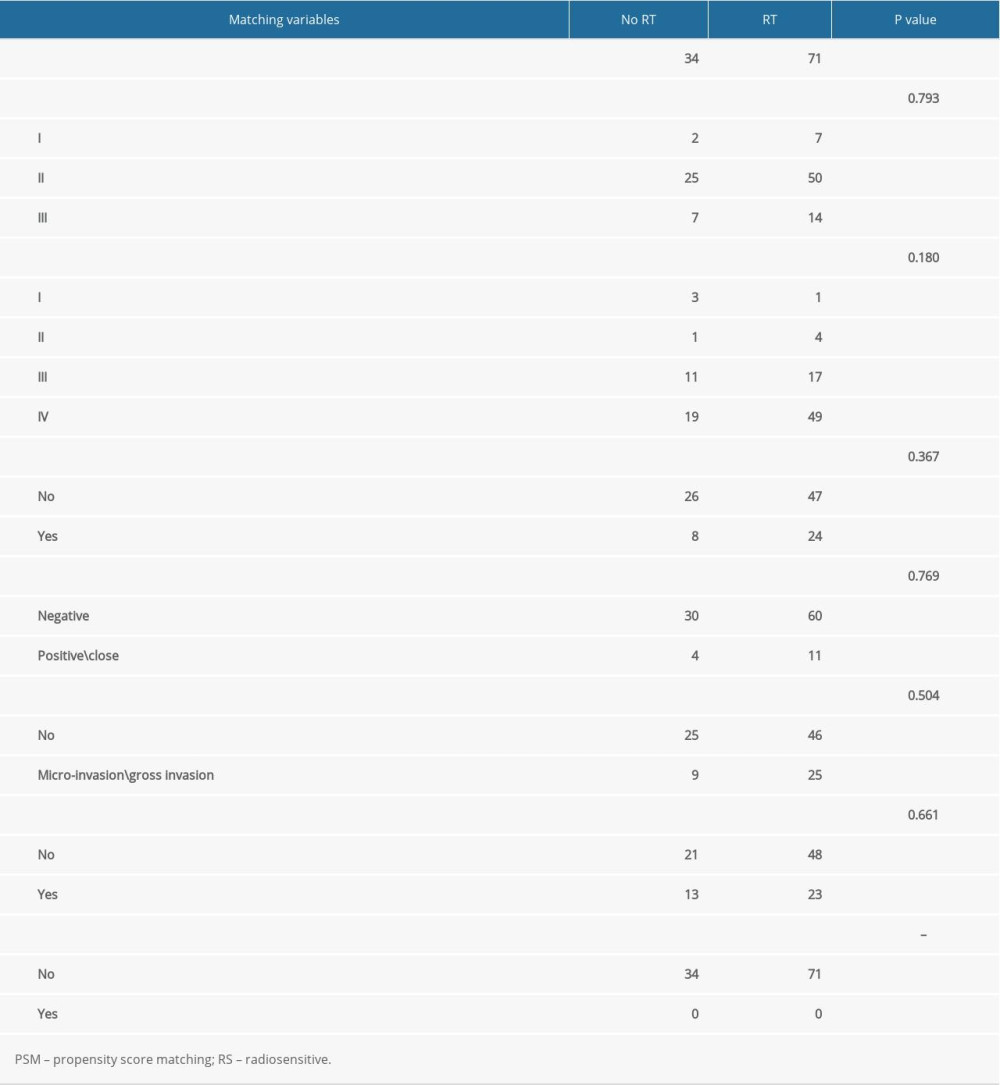 Table 3. Comparison of patients clinical and pathological characteristics in training and validation cohorts.
Table 3. Comparison of patients clinical and pathological characteristics in training and validation cohorts.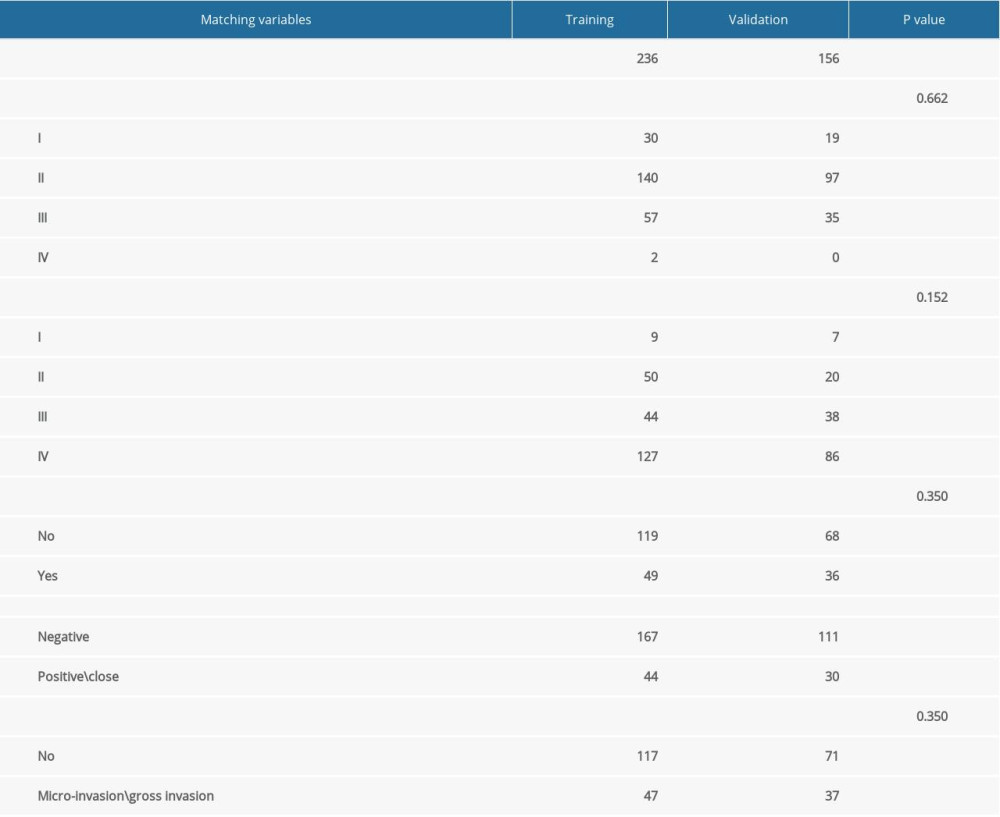 Table 4. Univariate and multivariate Cox regression analyses of interaction between RSS (as continues variable) and RT in training and validation cohort.
Table 4. Univariate and multivariate Cox regression analyses of interaction between RSS (as continues variable) and RT in training and validation cohort.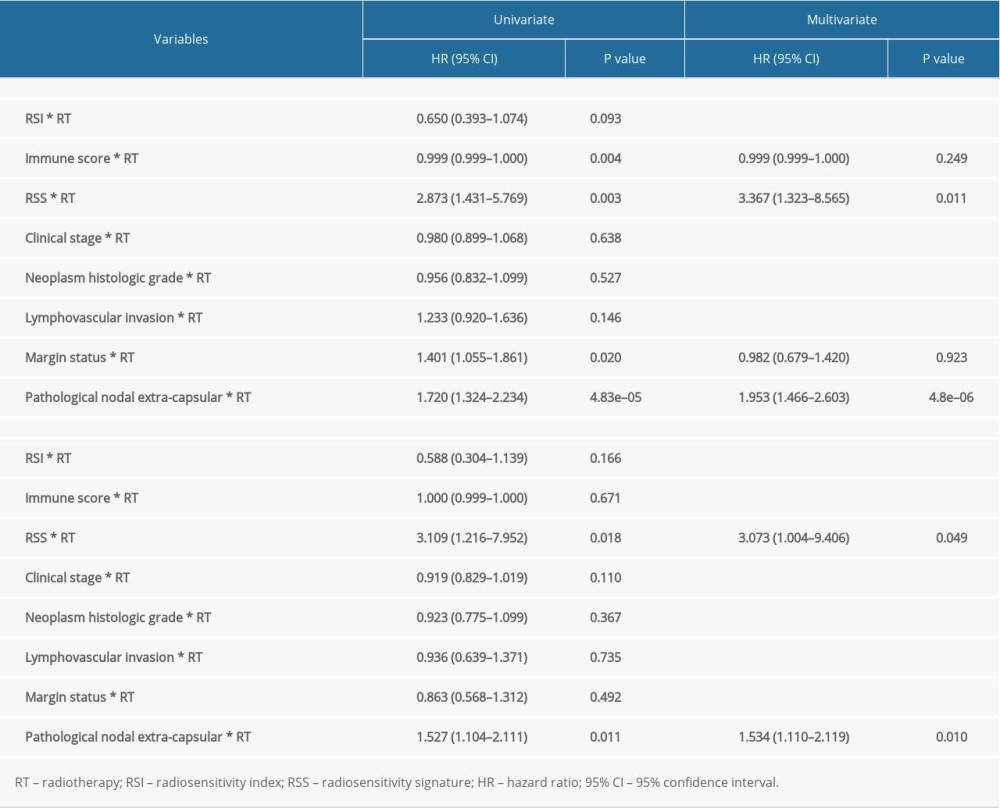
References
1. Argiris A, Karamouzis MV, Raben D, Ferris RL, Head and neck cancer: Lancet, 2008; 371; 1695-709
2. Johnson DE, Burtness B, Leemans CR, Head and neck squamous cell carcinoma: Nat Rev Dis Primers, 2020; 6; 92
3. Canning M, Guo G, Yu M, Heterogeneity of the head and neck squamous cell carcinoma immune landscape and its impact on immunotherapy: Front Cell Dev Biol, 2019; 7; 52
4. Plavc G, Jesenko T, Oražem M, Strojan P, Challenges in combining immunotherapy with radiotherapy in recurrent/metastatic head and neck cancer: Cancers (Basel), 2020; 12; 3197
5. Trotti A, Bellm LA, Epstein JB, Mucositis incidence, severity and associated outcomes in patients with head and neck cancer receiving radiotherapy with or without chemotherapy: A systematic literature review: Radiother Oncol, 2003; 66; 253-62
6. Rosenberg AJ, Vokes EE, Optimizing treatment de-escalation in head and neck cancer: Current and future perspectives: Oncologist, 2021; 26; 40-48
7. Chen AM, Daly ME, Farwell DG, Quality of life among long-term survivors of head and neck cancer treated by intensity-modulated radiotherapy: JAMA Otolaryngol Head Neck Surg, 2014; 140; 129-33
8. El-Deiry M, Funk GF, Nalwa SM, Long-term quality of life for surgical and nonsurgical treatment of head and neck cancer: Arch Otolaryngol Head Neck Surg, 2005; 131; 879-85
9. Eschrich SA, Pramana J, Zhang H, A gene expression model of intrinsic tumor radiosensitivity: Prediction of response and prognosis after chemoradiation: Int J Radiat Oncol Biol Phys, 2009; 75; 489-96
10. Caudell JJ, Torres-Roca JF, Gillies RJ, The future of personalised radiotherapy for head and neck cancer: Lancet Oncol, 2017; 18; e266-73
11. Ma J, Li R, Wang J, Characterization of a prognostic four-gene methylation signature associated with radiotherapy for head and neck squamous cell carcinoma: Mol Med Rep, 2019; 20; 622-32
12. You GR, Cheng AJ, Lee LY, Prognostic signature associated with radioresistance in head and neck cancer via transcriptomic and bioinformatic analyses: BMC Cancer, 2019; 19; 64
13. Cui Y, Li B, Pollom EL, Horst KC, Li R, Integrating radiosensitivity and immune gene signatures for predicting benefit of radiotherapy in breast cancer: Clin Cancer Res, 2018; 24; 4754-62
14. Chen DS, Mellman I, Oncology meets immunology: The cancer-immunity cycle: Immunity, 2013; 39; 1-10
15. Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2: Genome Biol, 2014; 15; 550
16. Ritchie ME, Phipson B, Wu D, limma powers differential expression analyses for RNA-sequencing and microarray studies: Nucleic Acids Res, 2015; 43; e47
17. Tian L, Alizadeh AA, Gentles AJ, Tibshirani R, A simple method for estimating interactions between a treatment and a large number of covariates: J Am Stat Assoc, 2014; 109; 1517-32
18. Shi LL, McMullen C, Vorwald K, Survival outcomes of patients with subglottic squamous cell carcinoma: A study of the National Cancer Database: Eur Arch Otorhinolaryngol, 2021 [Online ahead of print]
19. Chen AM, Chen LM, Vaughan A, Tobacco smoking during radiation therapy for head-and-neck cancer is associated with unfavorable outcome: Int J Radiat Oncol Biol Phys, 2011; 79; 414-19
20. Bentzen SM, Overgaard J, Patient-to-patient variability in the expression of radiation-induced normal tissue injury: Semin Radiat Oncol, 1994; 4; 68-80
21. Takeshima T, Chamoto K, Wakita D, Local radiation therapy inhibits tumor growth through the generation of tumor-specific CTL: Its potentiation by combination with Th1 cell therapy: Cancer Res, 2010; 70; 2697-706
22. Apetoh L, Ghiringhelli F, Tesniere A, Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy: Nat Med, 2007; 13; 1050-59
23. Lee Y, Auh SL, Wang Y, Therapeutic effects of ablative radiation on local tumor require CD8+ T cells: Changing strategies for cancer treatment: Blood, 2009; 114; 589-95
24. Kawaguchi T, Ono T, Sato F, CD8+ T cell infiltration predicts chemoradiosensitivity in nasopharyngeal or oropharyngeal cancer: Laryngoscope, 2021; 131; E1179-e89
25. Gurin D, Slavik M, Hermanova M, The tumor immune microenvironment and its implications for clinical outcome in patients with oropharyngeal squamous cell carcinoma: J Oral Pathol Med, 2020; 49; 886-96
26. Patel P, Sun L, Robbins Y, Enhancing direct cytotoxicity and response to immune checkpoint blockade following ionizing radiation with Wee1 kinase inhibition: Oncoimmunology, 2019; 8; e1638207
27. Plavc G, Strojan P, Combining radiotherapy and immunotherapy in definitive treatment of head and neck squamous cell carcinoma: Review of current clinical trials: Radiol Oncol, 2020; 54; 377-93
28. Qin Y, Zheng X, Gao W, Tumor microenvironment and immune-related therapies of head and neck squamous cell carcinoma: Mol Ther Oncolytics, 2021; 20; 342-51
Figures
 Figure 1. Schema of patient grouping. RSI – radiosensitivity index; RS – radiosensitive; RR – radioresistant; PSM – propensity score matching.
Figure 1. Schema of patient grouping. RSI – radiosensitivity index; RS – radiosensitive; RR – radioresistant; PSM – propensity score matching. Figure 2. Kaplan-Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the group of (A) RS (B) RR (C) immune (D) non-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy.
Figure 2. Kaplan-Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the group of (A) RS (B) RR (C) immune (D) non-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy. Figure 3. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the subgroup of (A) RS-immune (B) RS-non-immune (C) RR-immune (D) RR-non-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy.
Figure 3. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the subgroup of (A) RS-immune (B) RS-non-immune (C) RR-immune (D) RR-non-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy. Figure 4. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the matching group of (A) RS-immune (B) non-RS-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy.
Figure 4. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the matching group of (A) RS-immune (B) non-RS-immune. RS – radiosensitive; RR – radioresistant; RT – radiotherapy. Figure 5. Gene Ontology and KEGG pathway enrichment analysis of differentially expressed genes between the RS-immune group and non-RS-immune group. The top 10 GO biological process enrichment results (A) and top 10 KEGG enrichment results (B) for differentially expressed genes.
Figure 5. Gene Ontology and KEGG pathway enrichment analysis of differentially expressed genes between the RS-immune group and non-RS-immune group. The top 10 GO biological process enrichment results (A) and top 10 KEGG enrichment results (B) for differentially expressed genes. Figure 6. Signature established by L2 regularized (ridge) Cox regression model. (A) Parameter (Lambda2) selection by L2 regularized (ridge) Cox regression model adopted 10-fold cross-validation; (B) L2 regularized (ridge) Cox regression model coefficient profile plot of interaction between radiotherapy and genes selected by univariate Cox regression analyses. cvl – cross-validated likelihood; RT – radiotherapy.
Figure 6. Signature established by L2 regularized (ridge) Cox regression model. (A) Parameter (Lambda2) selection by L2 regularized (ridge) Cox regression model adopted 10-fold cross-validation; (B) L2 regularized (ridge) Cox regression model coefficient profile plot of interaction between radiotherapy and genes selected by univariate Cox regression analyses. cvl – cross-validated likelihood; RT – radiotherapy. Figure 7. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the group of (A) RSS-L (B) RSS-H in the training cohort and in the group of (C) RSS-L (D) RSS-H in validation cohort. The median values of the RSS score were used as the cut offs to define the high and low RSS groups in both training and validation cohorts. RRS-L – RSS low expression; RSS-H – RSS high expression; RT – radiotherapy.
Figure 7. Kaplan Meier survival analysis. Kaplan Meier plots were drawn to compare OS between patients receiving and not receiving RT in the group of (A) RSS-L (B) RSS-H in the training cohort and in the group of (C) RSS-L (D) RSS-H in validation cohort. The median values of the RSS score were used as the cut offs to define the high and low RSS groups in both training and validation cohorts. RRS-L – RSS low expression; RSS-H – RSS high expression; RT – radiotherapy. Tables
 Table 1. PSM result in RS-immune group.
Table 1. PSM result in RS-immune group. Table 2. PSM result in non-RS-immune group.
Table 2. PSM result in non-RS-immune group. Table 3. Comparison of patients clinical and pathological characteristics in training and validation cohorts.
Table 3. Comparison of patients clinical and pathological characteristics in training and validation cohorts. Table 4. Univariate and multivariate Cox regression analyses of interaction between RSS (as continues variable) and RT in training and validation cohort.
Table 4. Univariate and multivariate Cox regression analyses of interaction between RSS (as continues variable) and RT in training and validation cohort. Table 1. PSM result in RS-immune group.
Table 1. PSM result in RS-immune group. Table 2. PSM result in non-RS-immune group.
Table 2. PSM result in non-RS-immune group. Table 3. Comparison of patients clinical and pathological characteristics in training and validation cohorts.
Table 3. Comparison of patients clinical and pathological characteristics in training and validation cohorts. Table 4. Univariate and multivariate Cox regression analyses of interaction between RSS (as continues variable) and RT in training and validation cohort.
Table 4. Univariate and multivariate Cox regression analyses of interaction between RSS (as continues variable) and RT in training and validation cohort. In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952








