19 September 2020: Database Analysis
Prognostic Value of SLC4A4 and its Correlation with Immune Infiltration in Colon Adenocarcinoma
Xiaoli Chen1ABCDEF*, Jianing Chen2ABCDEF, Yan Feng1ABCDF, Wei Guan3ABCEFDOI: 10.12659/MSM.925016
Med Sci Monit 2020; 26:e925016
Abstract
BACKGROUND: SLC4A4 is differentially expressed in a variety of tumors, but its significance in colon adenocarcinoma has not been determined.
MATERIAL AND METHODS: Transcriptomes of two cohorts, GSE41258 and GSE32323, contained in The Cancer Genome Atlas (TCGA) were analysed to determine differences in SLC4A4 expression between tumor and normal tissue and their correlations with overall survival. The relationships between SLC4A4 expression and clinical characteristics were determined by COX regression analysis and logistic regression analysis, and correlations of SLC4A4 levels with tumor infiltrating immune cells (TIICs) and genes with high mutation frequency were evaluated by Pearson correlation analysis. Molecular functions and signaling pathways that might be affected by changes in SLC4A4 expression were determined by gene set enrichment analysis (GSEA). The overall distribution of TIICs was determined by two web servers: tumor immune estimation resource (TIMER) and CIBERSORT.
RESULTS: SLC4A4 expression was lower in colon adenocarcinoma than in normal colon tissue, suggesting that SLC4A4 was associated with poor prognosis. Reduced SLC4A4 expression was also associated with lymph node invasion and distant metastasis and was moderately correlated with increased expression of MUC4 and SMAD4, two genes with high mutation frequency in colon adenocarcinoma. GSEA indicated that changes in SLC4A4 expression affects several biological processes, including mismatch repair, base excision repair, and DNA replication. Eight TIICs in the tumor microenvironment differed significantly in groups with low and high expression of SLC4A4.
CONCLUSIONS: SLC4A4 may be a novel biomarker predicting prognosis in patients with colon adenocarcinoma. TIICs differed significantly in samples with higher and lower expression of SLC4A4.
Keywords: Sodium-Bicarbonate Symporters, Colonic Neoplasms, Databases, Factual, Neoplasm Proteins
Background
The prevalence of colorectal cancer is high, with estimates indicating that over 100,000 patients in the United States were diagnosed with colon cancer in 2019 [1]. Many risk factors affect the development and progression of colorectal cancer, including smoking, obesity, a low-fiber diet, and alcohol [2]. In addition, the incidence rate of colon cancer is higher in older than in younger people. Despite significant advances in individualized treatment, colon cancer is highly invasive and can metastasize to other organs, resulting in high mortality rates [3,4]. Moreover, colon cancer can bypass lymph nodes, making it more difficult to assess distant metastasis [5].
Most colorectal cancers are adenocarcinomas, with this disease showing significant heterogeneity. Cancer immunotherapy has been shown effective in colon cancer patients with DNA mismatch repair defects [6]. In addition, the immune microenvironment has been found to affect the occurrence and development of colon cancer [7,8]. Traditional categorical variables, such as tumor stage, make predicting the prognosis of patients difficult, as these categories lack individualization [9]. Several risk gene signatures have been shown effective in predicting the prognosis of patients with colon adenocarcinoma [10–12]. These risk genes were only used to build prognostic models, and their relationship with immune components in colon adenocarcinoma is unclear. Therefore, identifying new prognostic biomarkers and their association with immune characteristics is critical for better understanding of colon adenocarcinoma.
The solute carrier family 4 member 4 (SLC4A4) gene encodes a Na+/HCO3− cotransporter, which is mainly involved in the secretion and absorption of sodium bicarbonate. This process is highly important in maintaining the dynamic pH equilibrium within cells. SLC4A4 and other solute carrier family members have been found to contribute to tumorigenesis and development. For example, SLC4A4 or SLC4A9 have been reported to reduce intracellular pH and suppress tumor growth. The levels of expression of SLC4 and SLC26 family transporters have been shown to differ in colon cancer and normal colon tissue, suggesting that the dysregulation of bicarbonate transporters may be diagnostic of tumor development and may be a therapeutic target in cancer treatment [13]. MiR-223-3p decreases the expression of SLC4A4 in clear cell renal cell, enhancing tumor cell proliferation and metastasis [14]. SLC4A4 expression was also shown to be downregulated in thyroid cancer, suggesting diagnostic efficacy [15,16]. However, SLC4A4 is not downregulated in all tumors. For example, SLC4A4 expression was shown to be higher in mucinous epithelial ovarian cancer and chronic myeloid leukemia than in adjacent normal tissue, suggesting that the biological processes in which SLC4A4 is involved are tumor specific [17]. Gene signatures, including SLC4A4, can indicate the prognosis of patients with colorectal cancer [18–20]. Although SLC4A4 participates in the pathologenesis of colon cancers, thes association between its expression and the tumor immune microenvironment and classical mutant genes remain incompletely understood.
This study was performed to assess the association between SLC4A4 expression and patient prognosis by high-throughput sequencing of colon adenocarcinoma and normal colon samples acquired from The Cancer Genome Atlas (TCGA). Relative differences in SLC4A4 expression tissue and their correlations with overall survival were verified in two independent cohorts, GSE41258 and GSE32323. The associations between SLC4A4 expression and major clinical features and prognosis in patients with colon adenocarcinoma were analyzed statistically. The expression of SLC4A4 in other tumors was also examined to determine whether its expression has a general or essential effect on tumorigenesis. To better understand the relationship between SLC4A4 expression and gene mutations, the correlations between SLC4A4 expression and the 20 most frequently mutated genes in colon adenocarcinoma were calculated. Gene set enrichment analysis (GSEA) was performed based on gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. The associations between SLC4A4 expression and tumor-infiltrating immune cells (TIICs) were examined using of two web servers: tumor immune estimation resource (TIMER) and CIBERSORT. Finally, changes in SLC4A4 expression and their associations with patient prognosis were verified by gene expression profiling interactive analysis (GEPIA) and the Human Protein Atlas (HPA).
Material and Methods
CLINICAL INFORMATION AND EXPRESSION PROFILE:
Sequencing data on colon adenocarcinomas and corresponding clinical information in the TCGA were downloaded from the UCSC Xena database (https://xenabrowser.net/datapages/) [21], with org.Hs.eg.db (R package) used to annotate Ensembl ID. Data from some patients were excluded due to the absence of information on overall survival (OS), gender, age; tumor status, stage, or location; or lymph node or distant metastasis. Two independent cohorts (GSE41258 and GSE32323) were downloaded from the GEO database. GSE32323 contains 17 pairs of cancer and non-cancerous tissues, with gene expression measured by Affymetrix HG-U133 Plus 2.0 arrays. GSE41258 contains 390 samples, with genes measured by Affymetrix Human Genome U133A arrays. Primary tumor tissue from 153 patients with OS <120 months were selected and used in further analysis.
IMMUNE INFILTRATES ANALYSIS:
The association between SLC4A4 expression and the level of immune infiltration was assessed using the gene module in TIMER, a web server used for comprehensive assessment of levels of immune cell infiltration across different types of cancer [22]. TIMER uses a deconvolution method on high-throughput sequencing or microarray data to calculate the abundance of TIICs, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells. High-throughput sequencing data were evaluated using the CIBERSORT database, which assessed the compositional differences of TIICs between groups with high and low SLC4A4 expression. CIBERSORT uses the LM22 gene signature to classify 22 hematopoietic cell phenotypes into 6 categories, including myeloid cells, B cells, T cells, dendritic cells, macrophages and natural killer cells [23]. CIBERSORT normalized the estimates of the 22 TIICs in each sample to a sum of 1, allowing comparisons across samples and across immune cell types. The Mann-Whitney U test was performed to determine whether TIICs belong to different groups. In addition, Pearson’s correlation analysis between TIICs was performed to determine their possible associations. P<0.05 was defined as statistically significant.
GENE SET ENRICHMENT ANALYSIS:
GSEA was performed on normalized mRNA data using clusterprofiler (R package) [24]. The samples were separated into two groups based on the expression of SLC4A4, and the expression of other genes in the two groups analyzed using the R package, edgeR [25]. Fold changes (FCs) were calculated to show the magnitude of change in expression between the two groups. The gene list imported in into clusterprofiler was sorted in descending order by log2 FC. Permutations were set at 1000, the minimum size of each gene set for analysis at 10 and the maximum number size of genes annotated for testing at 1000. All entries in the GO database and the KEGG pathway were analyzed to investigate the biological functions in which SLC4A4 was involved. Entries with P<0.05 were considered statistically significant.
STATISTICAL ANALYSES:
Logistic regression analysis was performed to determine whether SLC4A4 can distinguish among patients with different clinical characteristics (age, gender, tumor status, lymph node, metastasis, stage); odds ratio (OR) and 95% confidence intervals (CIs) were calculated. Univariate COX regression analysis was performed to evaluate the associations of SLC4A4 and clinical features with OS by calculating hazard ratios (HRs) and 95% CIs. All statistical analyses were performed using R software (version 3.6.2), with p<0.05 considered statistically significant.
COMPREHENSIVE ANALYSIS:
Gene Expression Profiling Interactive Analysis (GEPIA) is a web server for transcriptome analysis [26] that includes 8,587 normal samples and 9,736 tumor samples from the GTEx and TCGA databases, with output provided by a standard processing pipeline. The association between SLC4A4 expression and survival in patients with colon adenocarcinoma was assessed by GEPIA. In addition, a boxplot was drawn to show where the disease state can be considered a variable to show differences in SLC4A4 expression among different tissue. Results were verified in two cohorts. The expression profiles in normal and tumor tissues were compared in GSE32323, and the association between SLC4A4 expression and OS in GSE41258.
TIMER includes genes with mutation frequency >10% in colon adenocarcinoma. To clarify the correlation between SLC4A4 expression and genes with high mutation frequency, the relationships between the expression of SLC4A4 and the 20 genes with highest mutation frequency were calculated. These 20 genes included APC, TP53, TTN, PIK3CA, KRAS, PTEN, ATM, MUC4, SMAD4, SYNE1, MUC16, FBXW7, BRAF, KIT, CSMD3, PTCH1, NEFH, NF1, RB1, and FAT4. Because cytokines are important components of the tumor microenvironment, the correlations between SLC4A4 expression and the expression of cytokine encoding genes, including IFNG, IL6, IL10, TGFB1, TGFB2, and TGFB3, were calculated. Pearson’s correlation coefficients for all of these associatins were calculated using the R package, Hmisc.
The human protein atlas (HPA;
Results
EXPRESSION PROFILE OF SLC4A4 AND ITS CORRELATION WITH OTHER GENES:
Differences in SLC4A4 expression were evaluated using the sequencing data obtained from TCGA. A comparison of 453 colon adenocarcinoma and 42 normal colon tissue samples showed that SLC4A4 expression was lower in the adenocarcinomas (Figure 1A), and that the density distribution of SLC4A4 expression differed in tumor tissue and normal tissue (Figure 1B). Pan-cancer analysis revealed that SLC4A4 expression was lower in 13 tumor types, including bladder urothelial carcinoma, breast invasive carcinoma, colon adenocarcinoma, esophageal carcinoma, head & neck squamous cell carcinoma, kidney chromophobe cell carcinoma, renal cell clear cell carcinoma, renal papillary cell carcinoma, hepatocellular carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, rectal adenocarcinoma and thyroid carcinoma, but was higher in cholangiocarcinoma and prostate adenocarcinoma. The correlations between SLC4A4 expression and the expression of genes encoding cytokines are presented in Figure 1C. The Pearson correlation coefficients of IFNG, IL6, IL10, TGFB1, TGFB2, and TGFB3 with SLC4A4 were 0.180 (P=2.01E-04), 0.117 (P=1.63E-02), 0.272 (P=1.21E-08), 0.219 (P=5.54E-06), 0.211 (P=1.19E-05) and 0.214 (P=8.77E-06), respectively. Figure 1D shows the correlations between SLC4A4 expression and the expression of genes with high mutation frequency in colon adenocarcinoma. The Pearson correlation coefficients of MUC4 and SMAD4 with SLC4A4 were 0.488 (P=7.95E-29) and 0.360 (P=1.81E-15), respectively.
RELATIONSHIP BETWEEN SLC4A4 EXPRESSION AND CLINICAL CHARACTERISTICS:
Figure 2A is a scatter plot showing the expression of SLC4A4, OS of patients with colon adenocarcinoma, and the average expression of SLC4A4 in patients who were dead or censored at the time of the last follow-up. The average expression of SLC4A4 was lower in patients who died than in the censored group, with the density peaks of these two groups being significantly different. Figure 2B shows that the expression of SLC4A4 was highly sensitive and specific in distinguishing colon adenocarcinoma from normal colon tissue (AUC=0.721). Univariate COX regression analysis showed that increased SLC4A4 expression was a protective factor (HR <1), being associated with increased OS. In addition, tumor status, lymph node disease, metastasis, and tumor stage wer associated with OS (Figure 3A). Logistic regression analysis comparing differences in SLC4A4 expression in patients with different clinical conditions found that the expression of SLC4A4 was significantly lower in groups with positive lymph node biopsy or distant metastasis than in groups without these conditions (Figure 3B). In addition, the average expression of SLC4A4 in stage IV colon adenocarcinoma was lower than in carcinoma tissue at other stages (Figure 3C). SLC4A4 expression was lower in stage IV tissue samples than in stage I, II, and III samples, with log2 FCs of −0.418 (P=5.30E-03), −0.585 (P=7.46E-06), and −0.348 (P=1.21E-02), respectively.
RESULTS OF FUNCTIONAL ENRICHMENT ANALYSIS:
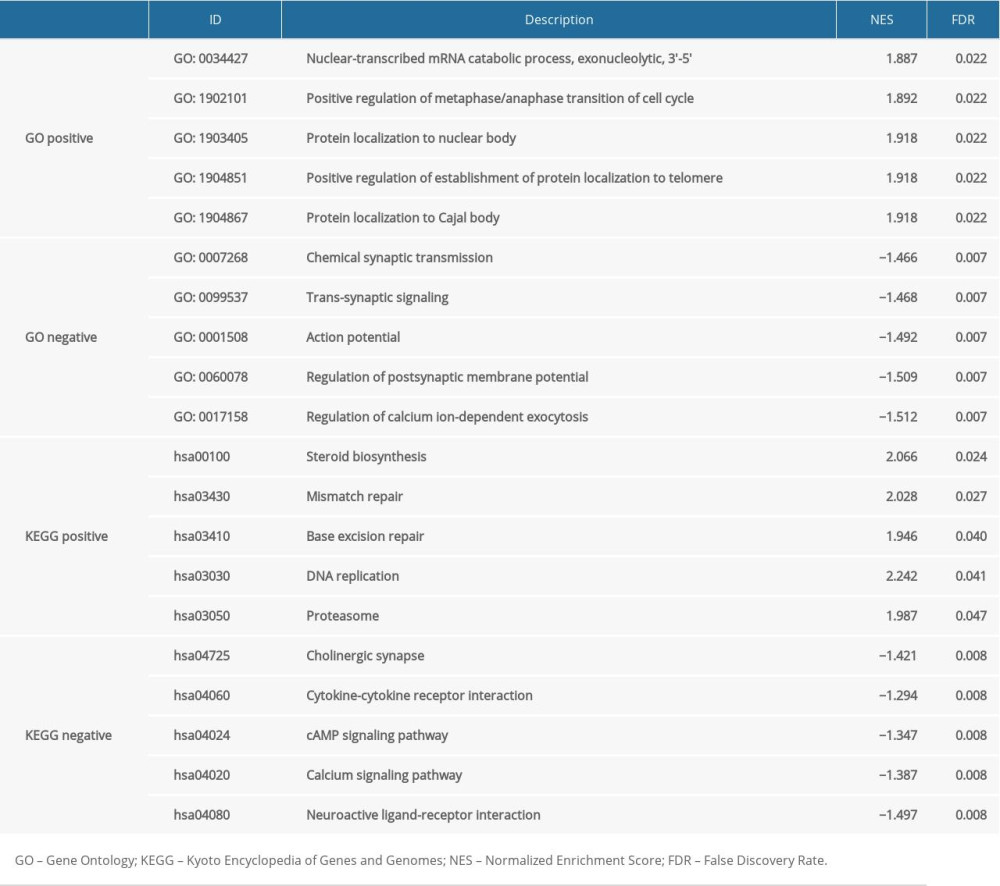
The influence of SLC4A4 expression on biological processes was analyzed by GO and KEGG pathways (Table 1). GSEA, a knowledge-based approach for interpreting genome-wide expression profiles, revealed a significantly different (FDR <0.05) functional pathway induced by decreased SLC4A4 expression [27]. Five GO entries were found to be positively correlated with decreased SLC4A4 expression, including nuclear-transcribed mRNA catabolic process, positive regulation of metaphase/anaphase transition of the cell cycle, protein localization to nuclear bodies, positive regulation of establishment of protein localization to telomeres and protein localization to Cajal bodies (Figure 4A). The five most significant entries that negatively correlated with decreased SLC4A4 expression were chemical synaptic transmission, trans-synaptic signaling, action potential, regulation of postsynaptic membrane potential and regulation of calcium ion-dependent exocytosis. In the KEGG pathway analysis, the five most positively correlated pathways with decreased SLC4A4 expression were steroid biosynthesis, mismatch repair, base excision repair, DNA replication and proteasome, whereas the five most negatively correlated pathways are cholinergic synapse, cytokine-cytokine receptor interaction, cAMP signaling pathway, calcium signaling pathway and neuroactive ligand-receptor interaction. The chronological sequence of these biological processes and tumorigenesis cannot be determined, so causality should be carefully evaluated. Some results of GSEA have been demonstrated to be associated with colon adenocarcinoma, whereas others require further research.
DNA mismatch repair is one of the main causes of colon adenocarcinoma [28]. Checkpoint inhibitors such as pembrolizumab and nivolumab after progression on irinotecan- or oxaliplatin-based therapies may benefit patients who exhibit DNA mismatch repair [29]. This imbalance in base excision repair may determine treatment response of colon adenocarcinomas and may be prognostic for survival [30,31].
CALCULATION AND EVALUATION OF TIICS:
Because tumor-infiltrating lymphocytes may be independently predictive of OS in cancer patients [32–34], the association between SLC4A4 expression and immune cell components in the tumor microenvironment were analyzed by TIMER. Immune cell populations that positively correlated with SLC4A4 expression in colon adenocarcinomas included B cells (P=9.66E-08), CD8+ T cells (P=3.53E-06), neutrophils (P=4.48E-06) and dendritic cells (P=2.89E-07). These results indicate that SLC4A4 performs an important function in the immune microenvironment of colon adenocarcinoma (Figure 4B).
To further analyze the association between SLC4A4 expression and immune cell components, all colon adenocarcinoma samples were divided into two groups based on median SLC4A4 expression. Samples with higher than median SLC4A4 concentrations were assigned to the high expression group and those with lower than median SLC4A4 concentrations to the low expression group. The association between SLC4A4 expression and TIICs was further clarified using an immune infiltration evaluation tool, CIBERSORT, to analyze the immune cell composition in each group, followed by statistical analyses using the Mann-Whitney U test (Figure 5A). CIBERSORT uses the LM22 gene signature to annotate the transcriptome and determine the composition of immune cells in samples. Because SLC4A4 expression is low in tumor samples, this group was defined as the high-risk group. The numbers of resting dendritic cells, activated mast cells, monocytes and resting memory CD4+ T cells were significantly lower in the high-risk group. In contrast, the numbers of some eosinophils, macrophages M0, resting mast cells and resting NK cells were significantly higher in the high-risk group. The infiltration levels of resting dendritic cells, eosinophil, monocytes and resting NK cells were too low (Figure 5B). Because activated and resting mast cells are two states of the same cells, the comprehensive changes are consistent. Calculations of the correlations among 22 types of immune cells showed weak or moderate correlations between the proportions of components (Figure 5C).
VERIFICATION OF SLC4A4 EXPRESSION CHARACTERISTICS:
GEPIA includes high-throughput sequencing data from tumor and normal tissue in the TCGA and GTEx databases. Because the GTEx database contains only normal tissues, GEPIA survival analysis was based on tumor samples from TCGA. Levels of SLC4A4 expression were divided into quartiles, with samples in the highest and lowest quartiles defined as the high and low expression groups, respectively. Kaplan-Meier analysis showed that the survival ratio was lower in the high expression group (Figure 6A), and a box plot showed that SLC4A4 expression differed in tumor and normal tissue samples (Figure 6B). Similar results were observed in the GSE32323 cohort (Figure 6C) and similar survival probability in the GSE41258 cohort (Figure 6D), suggesting that SLC4A4 is a protective factor in colon adenocarcinoma. Immunohistochemical analysis of the HPA database showed that SLC4A4 expression was lower in glandular cells of colon adenocarcinoma tissue than in normal colon gland cells (Figure 6E), confirming that SLC4A4 is a protective factor and its expression lower in colon adenocarcinoma than in normal colon tissue.
Discussion
Colon cancer is a universal public health problem and a major reason cause of cancer-related mortality [35]. Worldwide estimates indicate d that more than 2,200,000 individuals will be newly diagnosed with colon cancer in 2030, and that about 1,100,000 will die of this disease [36]. Early diagnosis requires the identification of appropriate molecular markers. The serum concentration of TRIM72 has been reported lower in patients with than without colon cancer, with TRIM72 being a better diagnostic indicator of colorectal cancer than CEA or CA199 alone [32]. When these three markers are used in combination, they are highly diagnostic, with an area under the curve of 0.928 [37].
The epithelial-mesenchymal transition is a marker of heterogeneity and pathological progression of colon cancer. CYB5R1 was found to be highly expressed in tumor cells undergoing the epithelial-mesenchymal transition, with CYB5R1 expression necessary for the invasive phenotype of tumor cells [38]. These studies indicate the significant clinical significance of identifying phenotypes through molecular features. To our knowledge, the current study is the first systematic analysis of the association between SLC4A4 expression and various pathological features of colon adenocarcinoma, including phenotype, tumor immune microenvironment, high mutation frequency gene profile, possible signaling pathways and prognosis.
We found that the expression of SLC4A4 was lower in colon adenocarcinoma than in normal colon tissue, a finding verified in the GEPIA and HPA databases. Because the expression of SLC4A4 is also significantly reduced in 12 other tumor types, SLC4A4 likely inhibits tumorigenesis through a ubiquitous mechanism. When SLC4A4 expression decreases, its protective effect is weakened, which indirectly promotes tumorigenesis. Logistic regression analysis showed that patients with low SLC4A4 expression were at higher risk for lymph node invasion and distant metastasis. SLC4A4 expression was lower in stage IV tumors than in other stages, suggesting that SLC4A4 may be involved in the occurrence of advanced events in colon tumorigenesis. SLC4A4 expression showed moderate correlations with MUC4 and SMAD4 mutations. Genetic variations in MUC4 affect the survival of patients with colon adenocarcinoma and are often accompanied by microsatellite instability [39,40]. Clonal loss of SMAD4 expression accumulates with disease progression in adenomas and promotes inflammation-driven colon cancer [41,42]. GSEA showed that SLC4A4A expression was associated with steroid biosynthesis, mismatch repair, base excision repair, DNA replication and the proteasome. Steroid biosynthesis is important in patients with hormone-dependent tumors, as anti-tumor drugs such as CYP17A1 inhibitors designed to suppress steroid biosynthesis are used in the treatment of prostate cancer [43,44]. Few studies to date, however, have analyzed whether steroid biosynthesis promotes or suppresses the development of colon adenocarcinoma, suggesting the need for additional research. Systemic mismatch repair deficiencies are usually manifested as microsatellite instability, resulting in the unique clinicopathological characteristics of individual colon adenocarcinomas [45]. Microsatellite instability can also predict tumor resistance to certain chemotherapeutic agents, such as immune checkpoint inhibitors [46]. Base excision repair is critical for removing endogenously damaged DNA bases, with failure to remove these bases leading to chromosomal instability and tumor development [47].
Tumor cells have three unique attributes, genomic instability, evasion of apoptosis, and continued proliferation. The DNA replication stress model proposes that genes that drive cell proliferation can induce DNA replication, which leads to genome instability and the selection of cells that can escape apoptosis [48]. The proteasome functions significantly in the occurrence, growth and invasion of tumor cells, with the proteasome considered a target for anti-cancer drugs [49]. Several newly discovered natural proteasome inhibitors and drugs found to function similarly have been used in clinical treatment [50,51]. The GSEA indicates the importance of the mechanism by which SLC4A4 participates in and affects mismatch repair, base excision repair, DNA replication and the proteasome.
The immunosuppressive mechanism in the tumor microenvironment is a protective barrier for tumor cells. Treatment of tumor tissue with various immunomodulators makes the tumor itself a source of antigens, which activate the dormant immune system, a promising treatment method known as in situ vaccination [52]. Because changes in immune components in the tumor microenvironment are of great significance, TIMER and CIBERSORT were applied to explore the association between SLC4A4 expression and immune cells in colon adenocarcinomas. The numbers of B cells, dendritic cells, CD8+ T cells, and neutrophils were found to positively correlate with SCL4A4 expression, indicating that the proportion of these cells was lower in tumor than in normal tissue. B cells in the tumor microenvironment can not only trigger humoral immunity and secrete inflammatory factors, but also recognize, process and present antigens. Moreover, B cells maintain positive interactions with other immune cells, including T cells and all immune cells expressing Fc receptors [53]. The prognostic significance of CD3+ T cells and CD8+ T cells is usually consistent with that of tumor infiltrating B cells, with these T cell populations also having a stronger prognostic significance when coexisting with tumor infiltrating B cells [54]. Dendritic cells activate CD8+ T cells through cross-presentation of exogenous antigens, a step considered key to activate CD8+ T cells against tumors [55,56]. These findings suggest that reductions in the numbers of dendritic cells and CD8+ T cells lead to a poor prognosis in patients with colon adenocarcinoma. Based on the CIBERSORT algorithm, we compared immune cells in tumors expressing high and low levels of SLC4A4, find that eight of the 22 types of immune cells differed significantly in these two groups, suggesting that SLC4A4 widely affects TIICs.
In summary, this study systematically analyzed the significance of SLC4A4 expression in colon adenocarcinoma, especially its association with tumor infiltrating immune cells. SLC4A4 may be a marker for the diagnosis of colon adenocarcinoma and a target for treatment, as well as making possible combinations of targeted therapy and immunotherapy in colon cancer.
Conclusions
SLC4A4 extensively affected tumor-infiltrating immune cells in colon adenocarcinoma tissues and was associated with tumor characteristics. SLC4A4 may be a new biomarker for the diagnosis and prognosis of patients with colon cancer.
Figures
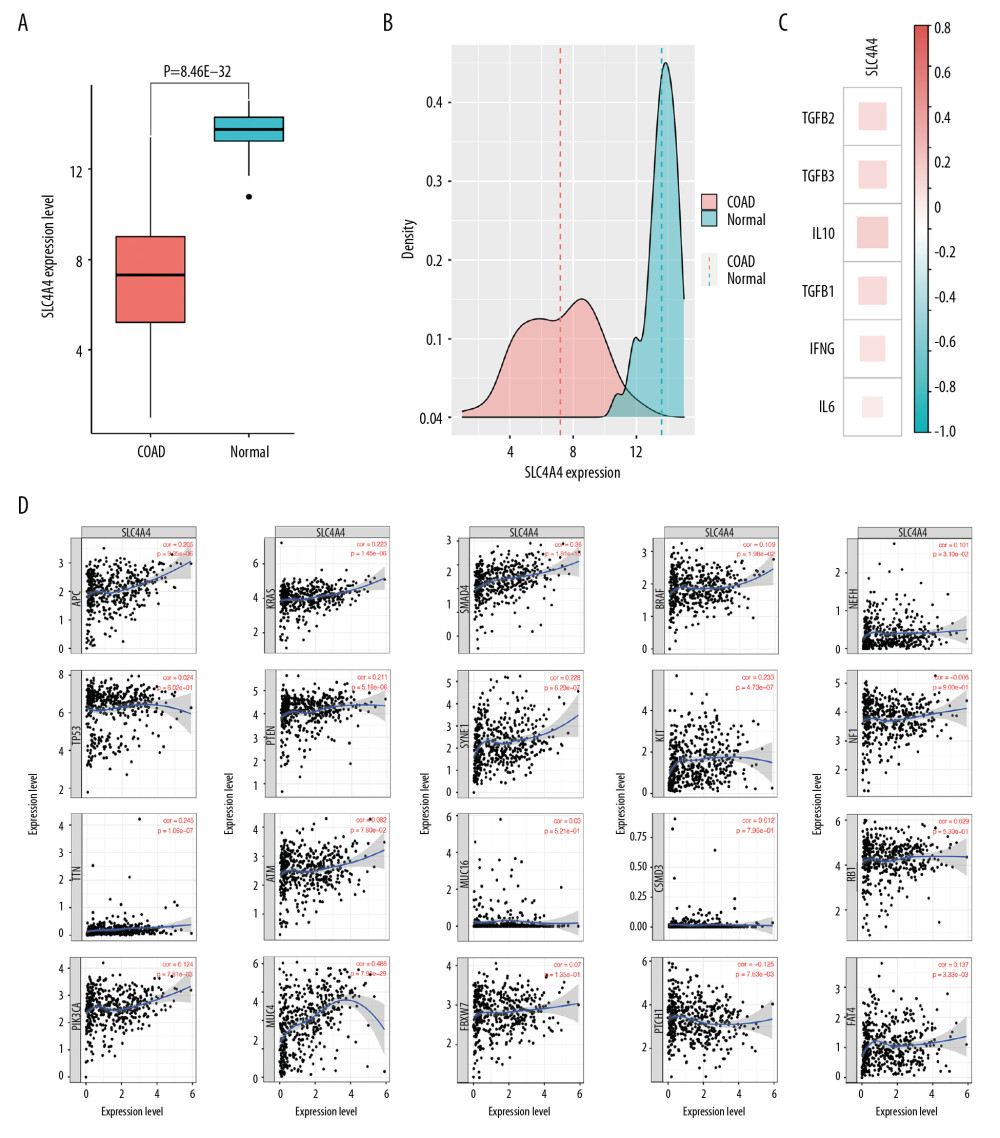 Figure 1. Expression of SLC4A4 in different cohorts and its correlation with the expression of other genes. (A) Expression of SLC4A4 in colon adenocarcinoma (COAD) and normal colon tissue. (B) Density plot of SLC4A4 expression in COAD and normal colon tissue. (C) Correlation between SLC4A4 expression and the expression of genes encoding cytokines. (D) Correlation between SLC4A4 expression and genes with high frequency of mutation in colon adenocarcinoma.
Figure 1. Expression of SLC4A4 in different cohorts and its correlation with the expression of other genes. (A) Expression of SLC4A4 in colon adenocarcinoma (COAD) and normal colon tissue. (B) Density plot of SLC4A4 expression in COAD and normal colon tissue. (C) Correlation between SLC4A4 expression and the expression of genes encoding cytokines. (D) Correlation between SLC4A4 expression and genes with high frequency of mutation in colon adenocarcinoma. 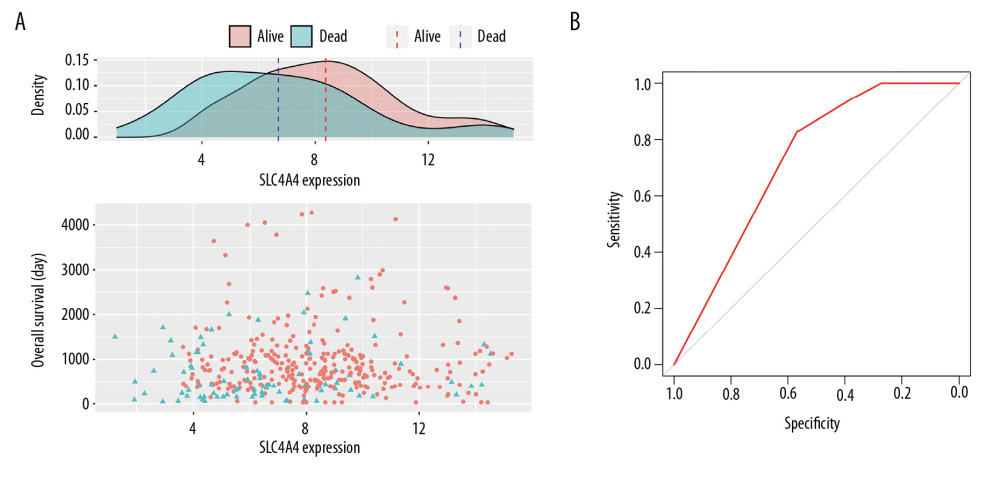 Figure 2. Relationship between SLC4A4 expression and overall survival. (A) Distribution of SLC4A4 expression and overall survival (OS). (B) Receiver operating characteristic (ROC) curve of SLC4A4 concentration distinguishing colon adenocarcinoma tissues from normal tissues.
Figure 2. Relationship between SLC4A4 expression and overall survival. (A) Distribution of SLC4A4 expression and overall survival (OS). (B) Receiver operating characteristic (ROC) curve of SLC4A4 concentration distinguishing colon adenocarcinoma tissues from normal tissues. 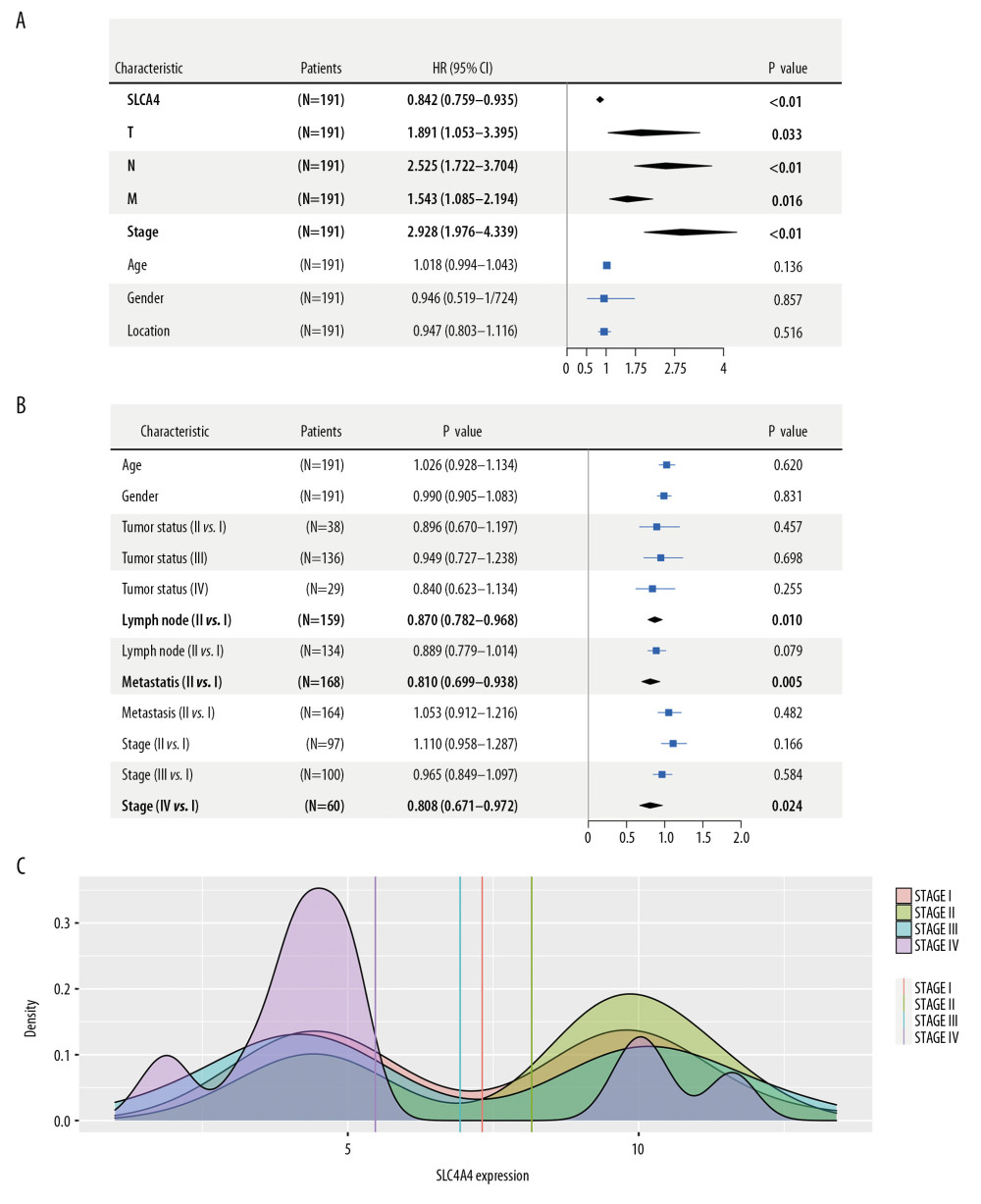 Figure 3. Association between the expression of SLC4A4 and clinical characteristics. (A) Forest plot of univariate COX regression analysis. T – tumor status; N – lymph node; M – metastasis. (B) Forest plot of logistic regression. (C) Density plot showing the expression of SLC4A4 at different stages of colon adenocarcinoma.
Figure 3. Association between the expression of SLC4A4 and clinical characteristics. (A) Forest plot of univariate COX regression analysis. T – tumor status; N – lymph node; M – metastasis. (B) Forest plot of logistic regression. (C) Density plot showing the expression of SLC4A4 at different stages of colon adenocarcinoma. 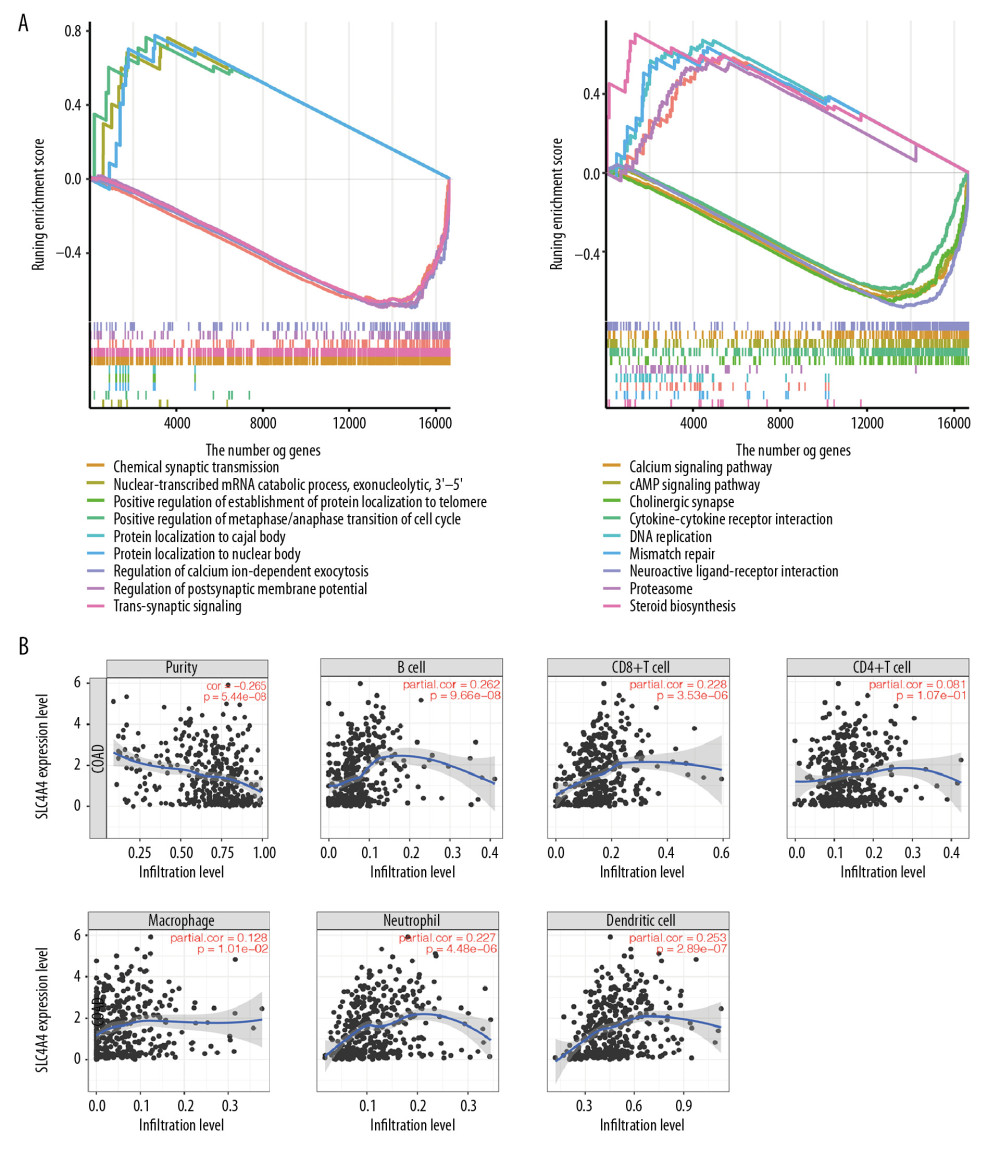 Figure 4. The results of GSEA and the correlation between SLC4A4 and TIICs. (A) The results of GSEA in the GO database (left) and the KEGG database (right). (B) Correlations between SLC4A4 expression and tumor-infiltrating immune cells.
Figure 4. The results of GSEA and the correlation between SLC4A4 and TIICs. (A) The results of GSEA in the GO database (left) and the KEGG database (right). (B) Correlations between SLC4A4 expression and tumor-infiltrating immune cells. 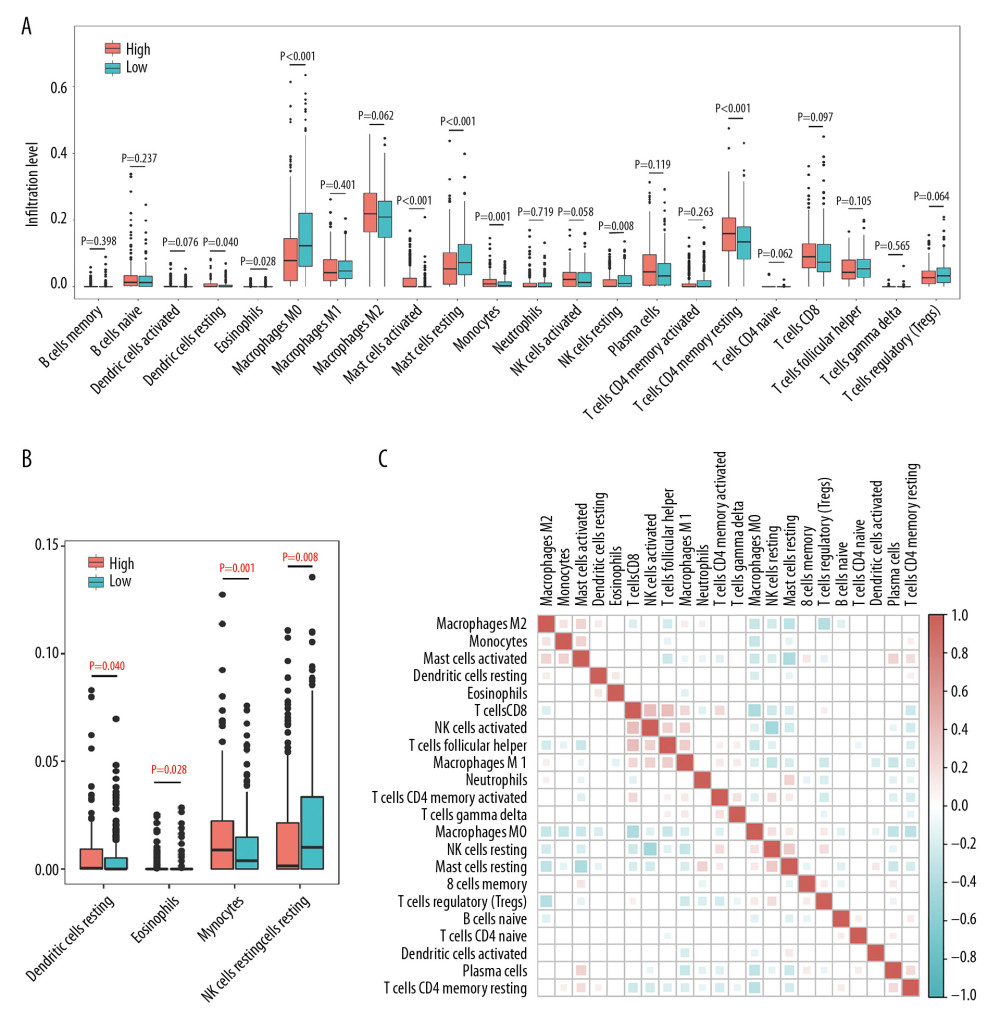 Figure 5. Differences between TIIC populations in samples with high and low SLC4A4 expression and correlations among TIICs. (A) Numbers of 22 types of immune cells in colon carcinoma tissues with high and low levels of SLC4A4 expression. (B) Four types of immune cells with low levels of infiltration. (C) Correlation among the numbers of 22 types of immune cells.
Figure 5. Differences between TIIC populations in samples with high and low SLC4A4 expression and correlations among TIICs. (A) Numbers of 22 types of immune cells in colon carcinoma tissues with high and low levels of SLC4A4 expression. (B) Four types of immune cells with low levels of infiltration. (C) Correlation among the numbers of 22 types of immune cells. 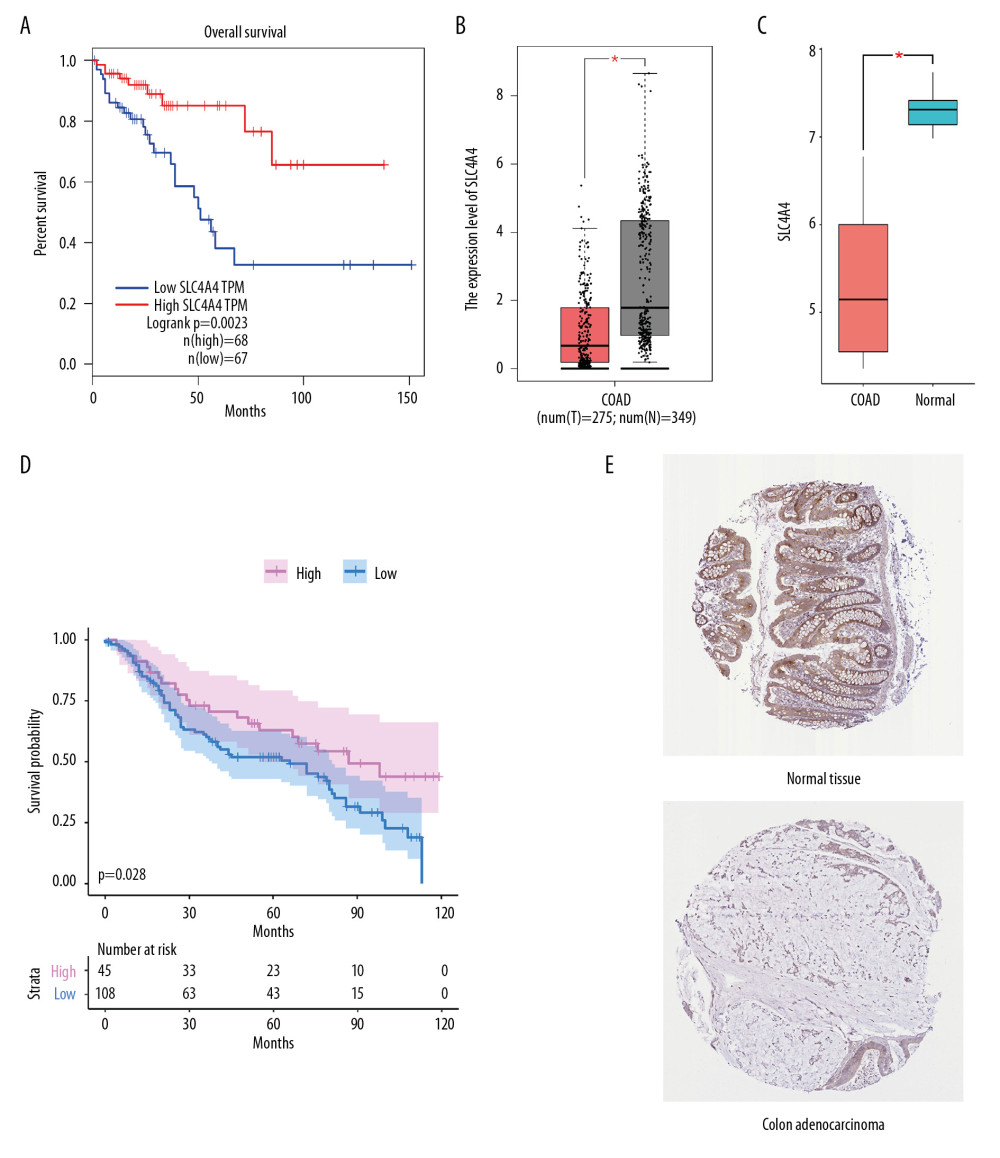 Figure 6. Validation from GEPIA and HPA. (A) Kaplan Meier analysis of the relationship between overall survival (OS) and SLC4A4 expression in GEPIA. (B, C) Levels of SLC4A4 expression in colon adenocarcinoma (COAD) and normal colon tissue in (B) GEPIA and (C) the GSE32323 cohort. (D) Kaplan Meier analysis of OS as a function of SLC4A4 expression in the GSE41258 cohort. (E) Immunohistochemical analysis of the expression of SLC4A4 protein in colon adenocarcinoma and normal colon tissue.
Figure 6. Validation from GEPIA and HPA. (A) Kaplan Meier analysis of the relationship between overall survival (OS) and SLC4A4 expression in GEPIA. (B, C) Levels of SLC4A4 expression in colon adenocarcinoma (COAD) and normal colon tissue in (B) GEPIA and (C) the GSE32323 cohort. (D) Kaplan Meier analysis of OS as a function of SLC4A4 expression in the GSE41258 cohort. (E) Immunohistochemical analysis of the expression of SLC4A4 protein in colon adenocarcinoma and normal colon tissue. References
1. Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019: Cancer J Clin, 2019; 69(1); 7-34
2. Islami F, Goding Sauer A, Miller KD, Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States: Cancer J Clin, 2018; 68(1); 31-54
3. Kalyan A, Kircher S, Shah H, Updates on immunotherapy for colorectal cancer: J Gastrointest Oncol, 2018; 9(1); 160-69
4. Sun Y, Zheng ZP, Li H: Mol Med Rep, 2016; 14(2); 1714-20
5. , Spreading colon cancer can bypass lymph nodes: Cancer Discov, 2017; 7; 924-25
6. Lazarus J, Maj T, Smith JJ, Spatial and phenotypic immune profiling of metastatic colon cancer: JCI Insight, 2018; 3(22); e121932
7. Llosa NJ, Cruise M, Tam A, The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints: Cancer Discov, 2015; 5(1); 43-51
8. Qiao G, Qin J, Kunda N, LIGHT elevation enhances immune eradication of colon cancer metastases: Cancer Res, 2017; 77(8); 1880-91
9. Vogelstein B, Papadopoulos N, Velculescu VE, Cancer genome landscapes: Science, 2013; 339(6127); 1546-58
10. Yang H, Liu H, Lin HC, Association of a novel seven-gene expression signature with the disease prognosis in colon cancer patients: Aging (Albany NY), 2019; 11(19); 8710-27
11. Sun D, Chen J, Liu L, Establishment of a 12-gene expression signature to predict colon cancer prognosis: Peer J, 2018; 6; e4942
12. Zuo S, Dai G, Ren X, Identification of a 6-gene signature predicting prognosis for colorectal cancer: Cancer Cell Int, 2019; 19; 6
13. McIntyre A, Hulikova A, Ledaki I, Disrupting hypoxia-induced bicarbonate transport acidifies tumor cells and suppresses tumor growth: Cancer Res, 2016; 76(13); 3744-55
14. Xiao W, Wang X, Wang T, Xing J, MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma: Aging (Albany NY), 2019; 11(2); 615-33
15. Gomez-Rueda H, Palacios-Corona R, Gutierrez-Hermosillo H, Trevino V, A robust biomarker of differential correlations improves the diagnosis of cytologically indeterminate thyroid cancers: Int J Mol Med, 2016; 37(5); 1355-62
16. Gałeza-Kulik M, Zebracka J, Szpak-Ulczok SExpression of selected genes involved in transport of ions in papillary thyroid carcinoma: Endokrynol Pol, 2006; 57(Suppl A); 26-31 [in Polish]
17. Gerber JM, Gucwa JL, Esopi D, Genome-wide comparison of the transcriptomes of highly enriched normal and chronic myeloid leukemia stem and progenitor cell populations: Oncotarget, 2013; 4(5); 715-28
18. Dai GP, Wang LP, Wen YQ, Identification of key genes for predicting colorectal cancer prognosis by integrated bioinformatics analysis: Oncol Lett, 2020; 19(1); 388-98
19. Gao X, Yang J, Identification of genes related to clinicopathological characteristics and prognosis of patients with colorectal cancer: DNA Cell Biol, 2020; 39(4); 690-99
20. Long NP, Park S, Anh NH, High-throughput omics and statistical learning integration for the discovery and validation of novel diagnostic signatures in colorectal cancer: Int J Mol Sci, 2019; 20(2); 296
21. Wang Z, Jensen MA, Zenklusen JC, A practical guide to The Cancer Genome Atlas (TCGA): Methods Mol Biol, 2016; 1418; 111-41
22. Li T, Fan J, Wang B, TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells: Cancer Res, 2017; 77(21); e108-10
23. Newman AM, Liu CL, Green MR, Robust enumeration of cell subsets from tissue expression profiles: Nat Methods, 2015; 12(5); 453-57
24. Yu G, Wang LG, Han Y, He QY, clusterProfiler: An R package for comparing biological themes among gene clusters: OMICS, 2012; 16(5); 284-87
25. Robinson MD, McCarthy DJ, Smyth GK, edgeR: A bioconductor package for differential expression analysis of digital gene expression data: Bioinformatics, 2010; 26(1); 139-40
26. Tang Z, Li C, Kang B, GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses: Nucleic Acids Res, 2017; 45; W98-102 W1
27. Subramanian A, Tamayo P, Mootha VK, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles: Proc Natl Acad Sci USA, 2005; 102(43); 15545-50
28. Recio-Boiles A, Waheed A, Cagir B: Cancer, colon, 2020, StatPearls, Treasure Island (FL)
29. Das S, Ciombor KK, Haraldsdottir S, Goldberg RM, Promising new agents for colorectal cancer: Curr Treat Options Oncol, 2018; 19(6); 29
30. Vodenkova S, Jiraskova K, Urbanova M, Base excision repair capacity as a determinant of prognosis and therapy response in colon cancer patients: DNA Repair (Amst), 2018; 72; 77-85
31. Leguisamo NM, Gloria HC, Kalil AN, Base excision repair imbalance in colorectal cancer has prognostic value and modulates response to chemotherapy: Oncotarget, 2017; 8(33); 54199-214
32. Djenidi F, Adam J, Goubar A, CD8+CD103+ tumor-infiltrating lymphocytes are tumor-specific tissue-resident memory T cells and a prognostic factor for survival in lung cancer patients: J Immunol, 2015; 194; 3475-86
33. Kong JC, Guerra GR, Pham T, Prognostic impact of tumor-infiltrating lymphocytes in primary and metastatic colorectal cancer: A systematic review and meta-analysis: Dis Colon Rectum, 2019; 62(4); 498-508
34. Stanton SE, Disis ML, Clinical significance of tumor-infiltrating lymphocytes in breast cancer: J Immunother Cancer, 2016; 4; 59
35. Chiang EPI, Tsai SY, Kuo YH, Caffeic acid derivatives inhibit the growth of colon cancer: Involvement of the PI3-K/Akt and AMPK signaling pathways: PLoS One, 2014; 9(6); e99631
36. Arnold M, Sierra MS, Laversanne M, Global patterns and trends in colorectal cancer incidence and mortality: Gut, 2017; 66(4); 683-91
37. Chen Z, Yin X, Li K, Serum levels of TRIM72 are lower among patients with colon cancer: Identification of a potential diagnostic marker: Tohoku J Exp Med, 2018; 245(1); 61-68
38. Woischke C, Blaj C, Schmidt EM, CYB5R1 links epithelial-mesenchymal transition and poor prognosis in colorectal cancer: Oncotarget, 2016; 7(21); 31350-60
39. Pastrello C, Santarosa M, Fornasarig M, MUC gene abnormalities in sporadic and hereditary mucinous colon cancers with microsatellite instability: Dis Markers, 2005; 21(3); 121-26
40. Lu S, Catalano C, Huhn S, Single nucleotide polymorphisms within MUC4 are associated with colorectal cancer survival: PLoS One, 2019; 14(5); e0216666
41. Means AL, Freeman TJ, Zhu J, Epithelial Smad4 deletion up-regulates inflammation and promotes inflammation-associated cancer: Cell Mol Gastroenterol Hepatol, 2018; 6(3); 257-76
42. Yan P, Klingbiel D, Saridaki Z, Reduced expression of SMAD4 is associated with poor survival in colon cancer: Clin Cancer Res, 2016; 22(12); 3037-47
43. Auchus ML, Auchus RJ, Human steroid biosynthesis for the oncologist: J Investig Med, 2012; 60(2); 495-503
44. Ferraldeschi R, Sharifi N, Auchus RJ, Attard G, Molecular pathways: Inhibiting steroid biosynthesis in prostate cancer: Clin Cancer Res, 2013; 19(13); 3353-59
45. Ryan E, Sheahan K, Creavin B, The current value of determining the mismatch repair status of colorectal cancer: A rationale for routine testing: Crit Rev Oncol Hematol, 2017; 116; 38-57
46. Baretti M, Le DT, DNA mismatch repair in cancer: Pharmacol Ther, 2018; 189; 45-62
47. Marsden CG, Dragon JA, Wallace SS, Sweasy JB, Base excision repair variants in cancer: Methods Enzymol, 2017; 591; 119-57
48. Macheret M, Halazonetis TD, DNA replication stress as a hallmark of cancer: Annu Rev Pathol, 2015; 10; 425-48
49. Chen Y, Zhang Y, Guo X, Proteasome dysregulation in human cancer: Implications for clinical therapies: Cancer Metastasis Rev, 2017; 36(4); 703-16
50. Ao N, Chen Q, Liu G, The small molecules targeting ubiquitin-proteasome system for cancer therapy: Comb Chem High Throughput Screen, 2017; 20(5); 403-13
51. Soave CL, Guerin T, Liu J, Dou QP, Targeting the ubiquitin-proteasome system for cancer treatment: discovering novel inhibitors from nature and drug repurposing: Cancer Metastasis Rev, 2017; 36(4); 717-36
52. Locy H, de Mey S, de Mey W, Immunomodulation of the tumor microenvironment: Turn foe into friend: Front Immunol, 2018; 9; 2909
53. Tsou P, Katayama H, Ostrin EJ, Hanash SM, The emerging role of B cells in tumor immunity: Cancer Res, 2016; 76(19); 5597-601
54. Wouters MCA, Nelson BH, Prognostic significance of tumor-infiltrating B cells and plasma cells in human cancer: Clin Cancer Res, 2018; 24(24); 6125-35
55. Fu C, Jiang A, Dendritic cells and CD8 T cell immunity in tumor microenvironment: Front Immunol, 2018; 9; 3059
56. Veglia F, Gabrilovich DI, Dendritic cells in cancer: The role revisited: Curr Opin Immunol, 2017; 45; 43-51
Figures
 Figure 1. Expression of SLC4A4 in different cohorts and its correlation with the expression of other genes. (A) Expression of SLC4A4 in colon adenocarcinoma (COAD) and normal colon tissue. (B) Density plot of SLC4A4 expression in COAD and normal colon tissue. (C) Correlation between SLC4A4 expression and the expression of genes encoding cytokines. (D) Correlation between SLC4A4 expression and genes with high frequency of mutation in colon adenocarcinoma.
Figure 1. Expression of SLC4A4 in different cohorts and its correlation with the expression of other genes. (A) Expression of SLC4A4 in colon adenocarcinoma (COAD) and normal colon tissue. (B) Density plot of SLC4A4 expression in COAD and normal colon tissue. (C) Correlation between SLC4A4 expression and the expression of genes encoding cytokines. (D) Correlation between SLC4A4 expression and genes with high frequency of mutation in colon adenocarcinoma. Figure 2. Relationship between SLC4A4 expression and overall survival. (A) Distribution of SLC4A4 expression and overall survival (OS). (B) Receiver operating characteristic (ROC) curve of SLC4A4 concentration distinguishing colon adenocarcinoma tissues from normal tissues.
Figure 2. Relationship between SLC4A4 expression and overall survival. (A) Distribution of SLC4A4 expression and overall survival (OS). (B) Receiver operating characteristic (ROC) curve of SLC4A4 concentration distinguishing colon adenocarcinoma tissues from normal tissues. Figure 3. Association between the expression of SLC4A4 and clinical characteristics. (A) Forest plot of univariate COX regression analysis. T – tumor status; N – lymph node; M – metastasis. (B) Forest plot of logistic regression. (C) Density plot showing the expression of SLC4A4 at different stages of colon adenocarcinoma.
Figure 3. Association between the expression of SLC4A4 and clinical characteristics. (A) Forest plot of univariate COX regression analysis. T – tumor status; N – lymph node; M – metastasis. (B) Forest plot of logistic regression. (C) Density plot showing the expression of SLC4A4 at different stages of colon adenocarcinoma. Figure 4. The results of GSEA and the correlation between SLC4A4 and TIICs. (A) The results of GSEA in the GO database (left) and the KEGG database (right). (B) Correlations between SLC4A4 expression and tumor-infiltrating immune cells.
Figure 4. The results of GSEA and the correlation between SLC4A4 and TIICs. (A) The results of GSEA in the GO database (left) and the KEGG database (right). (B) Correlations between SLC4A4 expression and tumor-infiltrating immune cells. Figure 5. Differences between TIIC populations in samples with high and low SLC4A4 expression and correlations among TIICs. (A) Numbers of 22 types of immune cells in colon carcinoma tissues with high and low levels of SLC4A4 expression. (B) Four types of immune cells with low levels of infiltration. (C) Correlation among the numbers of 22 types of immune cells.
Figure 5. Differences between TIIC populations in samples with high and low SLC4A4 expression and correlations among TIICs. (A) Numbers of 22 types of immune cells in colon carcinoma tissues with high and low levels of SLC4A4 expression. (B) Four types of immune cells with low levels of infiltration. (C) Correlation among the numbers of 22 types of immune cells. Figure 6. Validation from GEPIA and HPA. (A) Kaplan Meier analysis of the relationship between overall survival (OS) and SLC4A4 expression in GEPIA. (B, C) Levels of SLC4A4 expression in colon adenocarcinoma (COAD) and normal colon tissue in (B) GEPIA and (C) the GSE32323 cohort. (D) Kaplan Meier analysis of OS as a function of SLC4A4 expression in the GSE41258 cohort. (E) Immunohistochemical analysis of the expression of SLC4A4 protein in colon adenocarcinoma and normal colon tissue.
Figure 6. Validation from GEPIA and HPA. (A) Kaplan Meier analysis of the relationship between overall survival (OS) and SLC4A4 expression in GEPIA. (B, C) Levels of SLC4A4 expression in colon adenocarcinoma (COAD) and normal colon tissue in (B) GEPIA and (C) the GSE32323 cohort. (D) Kaplan Meier analysis of OS as a function of SLC4A4 expression in the GSE41258 cohort. (E) Immunohistochemical analysis of the expression of SLC4A4 protein in colon adenocarcinoma and normal colon tissue. In Press
06 Mar 2024 : Clinical Research
Prevalence and Variation of Medical Comorbidities in Oral Surgery Patients: A Retrospective Study at Jazan ...Med Sci Monit In Press; DOI: 10.12659/MSM.943884
08 Mar 2024 : Clinical Research
Evaluation of Foot Structure in Preschool Children Based on Body MassMed Sci Monit In Press; DOI: 10.12659/MSM.943765
15 Apr 2024 : Laboratory Research
The Role of Copper-Induced M2 Macrophage Polarization in Protecting Cartilage Matrix in OsteoarthritisMed Sci Monit In Press; DOI: 10.12659/MSM.943738
07 Mar 2024 : Clinical Research
Knowledge of and Attitudes Toward Clinical Trials: A Questionnaire-Based Study of 179 Male Third- and Fourt...Med Sci Monit In Press; DOI: 10.12659/MSM.943468
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952