23 October 2020: Animal Study
Long Non-Coding RNA Nuclear-Enriched Abundant Transcript 1 (NEAT1) Represses Proliferation of Trophoblast Cells in Rats with Preeclampsia via the MicroRNA-373/FLT1 Axis
Lingling Teng1ABEF, Pingping Liu2ABDE, Xiao Song3BCE, Hui Wang4CEF, Jing Sun5BDE, Zhongxia Yin6CDEF*DOI: 10.12659/MSM.927305
Med Sci Monit 2020; 26:e927305
Abstract
BACKGROUND: Preeclampsia (PE) remains one of the primary causes of maternal morbidity and mortality worldwide. This study was designed to investigate the relevance of long non-coding RNA (lncRNA) nuclear-enriched abundant transcript 1 (NEAT1) and downstream molecules in trophoblast cell proliferation and apoptosis.
MATERIAL AND METHODS: NEAT1 expression in the placental tissues of rats with PE was analyzed by reverse transcriptionquantitative polymerase chain reaction. The role of NEAT in trophoblast cell proliferation, migration, invasion, and apoptosis was assessed by transfecting pcDNA-NEAT1 and siRNA-NEAT1 into trophoblast cells. The microRNA (miRNA) binding to NEAT1 and the genes targeted by the screened miRNAs were predicted by Starbase, and the mechanism of action of NEAT1 in PE was further investigated.
RESULTS: The expression of NEAT1 lncRNA was markedly higher in placental samples of PE than control rats. Ectopic expression of NEAT1 repressed trophoblast cell proliferation, migration, invasion, and colony formation, but facilitated cell apoptosis, whereas NEAT1 downregulation resulted in the opposite effects. NEAT1 was found to act as a molecular sponge for miR-373, regulating Fms-like tyrosine kinase-1 (FLT-1) to modulate PE development.
CONCLUSIONS: NEAT1 may contribute to PE development by regulating trophoblast cell proliferation and apoptosis. These findings may provide a new perspective for understanding the etiology and pathogenesis of PE.
Keywords: Pre-Eclampsia, Pregnancy, Trophoblasts, Vascular Endothelial Growth Factor Receptor-1
Background
Preeclampsia (PE) is a serious complication of pregnancy, characterized by hypertension, including a systolic blood pressure (SBP) of ≥140 mmHg and/or a diastolic blood pressure (DBP) of ≥90 mmHg, and proteinuria of ≥300 mg/day, leading to maternal and fetal morbidity and mortality [1]. In developed countries, postponement of childbearing increases the risks associated with PE, as delayed childbearing correlates with obesity and vascular diseases [2]. PE is also characterized by placental dysfunction, which is closely associated with the abnormal invasion and remodeling of maternal uterine arteries by trophoblasts [3]. Determining the molecular mechanisms associated with inadequate trophoblastic invasion and growth may help in understanding the pathogenesis of PE [4].
Long non-coding RNAs (lncRNAs), defined as RNAs >200 nucleotides in length with no protein-encoding potential, can regulate transcription within the nucleus and modulate mRNA stability, translation and post-translational alterations in the cytoplasm [5]. Moreover, several lncRNAs, including LOC391533, LOC284100, and CEACAMP8, have been reported to be associated with the pathogenesis of PE [6]. The expression of nuclear-enriched abundant transcript-1 (NEAT1) lncRNA at term was observed to be higher in placentas with than without intrauterine growth restriction [7], indicating that NEAT1 is associated with pregnancy. Nevertheless, the involvement of NEAT1 in PE remains unclear.
In addition, numerous microRNA (miRNA)-binding sites have been identified on a broad spectrum of RNA transcripts, suggesting that all RNAs with miRNA-binding sites have the ability to communicate with and modulate each other by competing for shared miRNAs, thereby acting as competing endogenous RNAs (ceRNAs) [8]. For example, the lncRNA HULC, which is enriched under conditions of oxidative stress, was found to mediate cholangiocarcinoma cell migration and invasion by interacting with CXCR4 via ceRNA-associated mechanisms of competing for miR-372/miR-373 [9]. miR-373, identified as a human embryonic stem cell-specific miRNA, was found to regulate cell proliferation, migration, invasion, and apoptosis [10], and was predicted to bind to NEAT1. Furthermore, Fms-like tyrosine kinase-1 (FLT-1), an antiangiogenic factor in the placenta, has been reported elevated in PE, leading to glomerular endothelial injury, hypertension, and proteinuria [11]. FLT-1 mRNA contains miR-373 binding sites, suggesting that FLT-1 is a putative target of miR-373 in PE. Taken together, these findings suggested that the NEAT1/miR-373/FLT-1 axis may modulate the proliferation, migration, invasion, and apoptosis of trophoblast cells in PE. Using a rat model of PE, this study examined the associations of the NEAT1/miR-373/FLT-1 axis with the biological activities of trophoblast cells, as well as the role of this axis in the exact mechanisms associated with the pathological development PE.
Material and Methods
RAT PE ANIMAL MODEL:
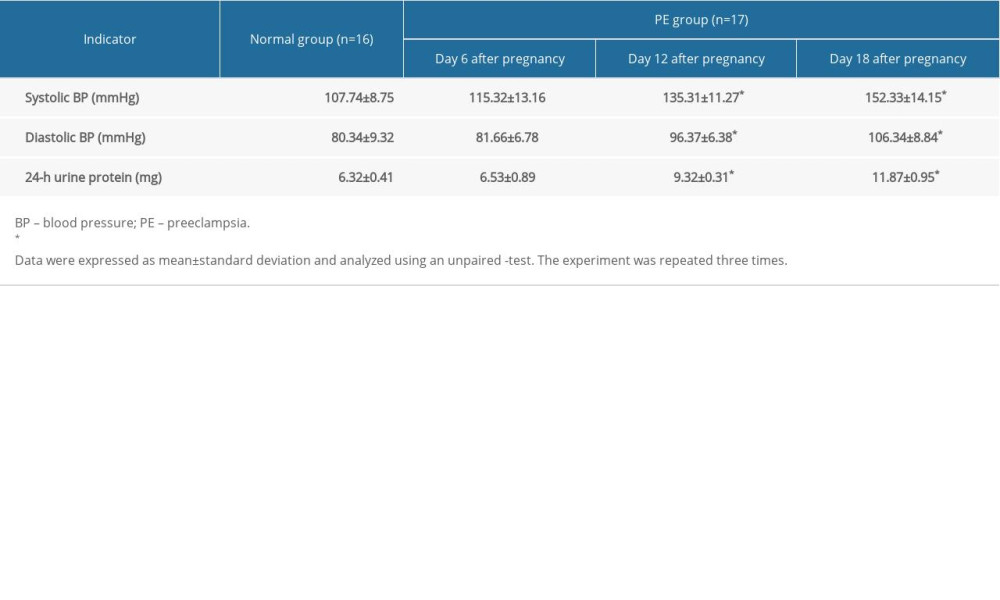
All animal protocols followed the guidelines of the Institutional Animal Care and Use Committee of Liaocheng Second People’s Hospital and were approved by the same committee (approval number: 20190014). Sixty Sprague-Dawley rats (40 females and 20 males, aged 2–3 months and weighing 200–220 g) were obtained from the laboratory animal center of Liaocheng Second People’s Hospital. Male and female rats were housed at a 1: 2 ratio, and pregnancy was established by the presence of spermatozoa in the vaginal smear, which was defined as day 0. The 33 rats with confirmed pregnancies were randomized to a PE (n=17) and a control (n=16) group on days 6 to 10 of pregnancy. PE was induced in pregnant rats as previously described [12] by continuous subcutaneous administration of 100 mg/kg/day NG-nitro-L-arginine methyl ester (D0819; Sigma-Aldrich Chemical Company, St. Louis, MO, USA); normal rats were administered the same volume of saline. Blood pressure (BP) and urine protein concentration were measured on days 6, 12, and 18 of pregnancy. On day 21, each rat was intraabdominally injected with 250 mg/kg pentobarbital sodium, with euthanasia confirmed successful by the absence of a heartbeat and a blinking reflex and the lack of spontaneous breathing for 2–3 min. The uterine placental tissues were harvested following cesarean section.
BIOCHEMICAL INDICATORS:
BP was measured at 8: 00 am daily after a 2-h fast and after 20 min in a thermostat preheated to 37°C. When heart rate had stabilized, BP was measured using a Coda noninvasive BP system (Kent Scientific Corporation, Torrington, CT, USA) and a tail-cuff device clamp.
Rats were housed individually in metabolic cages and 24-h urine samples were collected and centrifuged for 10 min at 717×g at 4°C to remove precipitates. Biuret reagent (HPBIO-R1253, Pengpai Biotech Company, Shanghai, China) was added to each sample, and urinary protein concentrations were measured using an automatic biochemical analyzer (AU5800; Beckman Coulter Inc., La Brea, CA, USA).
ISOLATION, TREATMENT, AND IDENTIFICATION OF PLACENTAL TROPHOBLAST CELLS:
Trophoblast cells were isolated and identified as described previously [13]. Briefly, following euthanasia, placentas were quickly removed from rats by opening the abdominal cavity. A small piece of tissue from the central maternal surface of the placenta with few blood vessels was immediately removed under direct vision, rinsed with saline solution, and sectioned into 1×1 mm pieces. These pieces were incubated with 0.25% trypsin (Beijing Solarbio Life Sciences Co., Ltd., Beijing, China) and 0.1% collagenase (Beijing Solarbio) overnight at 37°C, with the reaction terminated by addition of serum. The suspension was centrifuged at 1200×g for 30 min at 4°C. The collected trophoblasts were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.), 100 U/mL penicillin and 100 μg/mL streptomycin, at 37°C in a 5% CO2 incubator.
Trophoblast cells were seeded in 24-well culture plates and incubated for 48 h at 37°C. The cells were fixed in paraformaldehyde for 15 min at room temperature (20°C) and rinsed with phosphate-buffered saline (PBS) supplemented with 0.2% Triton X-100. After blocking with 1% rabbit serum for 30 min at room temperature, the cells were incubated with rabbit anti-mouse cytokeratin 7 (1: 8000, ab181598, Abcam Inc., Cambridge, UK) and vimentin (1: 500, ab92547, Abcam) antibodies overnight at 4°C and with Alexa Fluor® 488-conjugated secondary goat anti-rabbit IgG antibody (1: 200, ab15007, Abcam) for 2 h at 4°C. Following counter-staining with 4′,6-diamidino-2-phenylindole (DAPI) for 1 h, the cells were viewed with an inversion fluorescence microscope (IX71; Olympus Optical Co., Ltd., Tokyo, Japan) to detect the fluorescence and capture images. The proportion of stained cells was evaluated using Image-Pro Plus software (7.0; Media Cybernetics, Bethesda, MD, USA).
:
Following incubation with 4% formaldehyde for 15 min, trophoblast cells were treated with pepsin and ethanol. The dried trophoblast cells were mixed with hybridization buffer containing FISH probe NEAT1 (Guangzhou RiboBio Co., Ltd., Guangzhou, Guangdong, China) for 2 min at 80°C. After dehydration, the slices were counter-stained with DAPI, and the images were captured with a confocal microscopy (Leica Microsystems GmbH, Wetzlar, Germany).
SUBCELLULAR FRACTIONATION:
The nuclear and cytoplasmic fractions of trophoblast cells were separated using a PARIS kit (Life Technologies). Briefly, the cells were rinsed twice with precooled PBS and lysed in a mixture of 0.1% NP40, 10 mM ribonucleoside vanadyl complex (New England Biolabs, Ipswich, MA, USA) and proteinase inhibitors (Roche Diagnostics, Co., Ltd., Rotkreuz, Switzerland). After centrifugation, the lysates were washed 5 times with PBS and centrifuged to yield cytoplasmic RNA and nuclear RNA fractions.
RT-QPCR:
Total RNA was isolated from cells and tissues with TRIzol® reagent (Thermo Fisher Scientific Inc., Waltham, MA, USA), followed by synthesis of first chain cDNA using a PrimerScript RT kit (TaKaRa, Kyoto, Japan). RT-qPCR was performed using LightCycler 480SYBR Green I Master mixture (Roche Diagnostics) and primers for NEAT1
Levels of miRNA and mRNA were normalized relative to the levels of U6 and GAPDH, respectively, and calculated using the 2−ΔΔCt method.
CELL TRANSFECTION:
Vectors for the overexpression of NEAT1 (pcDNA-NEAT1) and FLT1 (pcDNA-FLT1), the miR-373 mimic/inhibitor and their respective negative controls (pcDNA, NC mimic and NC inhibitor) were purchased from RiboBio. Small interfering RNAs (siRNAs) targeting NEAT1 and FLT1 (si-NEAT1, si-FLT1) and control si-NC were obtained from GenePharma (Shanghai, China). Trophoblast cells were transfected with these vectors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
5-ETHYNYL-2′-DEOXYURIDINE (EDU) PROLIFERATION LABELING:
Trophoblast cells seeded onto 96-well plates were incubated with 50 μM 5-ethynyl-2′-deoxyuridine
PROLIFERATION ASSAY:
Cells, at a density of 2×104 cells per well, were plated in 96-well plates and cultured for 24 h. To each well was added 15 μL cell counting kit-8 (CCK-8) reagent, and the cells incubated for 2 h at 37°C. The cells were washed and incubated for 24, 48, and 72 h. The optical density (OD) at 490 nm was assessed using a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA).
For colony formation assays, cells in logarithmic phase were detached with trypsin (Gibco, Carlsbad, CA, USA) and resuspended in DMEM supplemented with 10% FBS. The cells were plated onto petri dishes at a density of 500 cells per dish and cultured at 37°C until colonies were visible. The medium was removed, and the cells were fixed with methanol and stained with crystal violet (Gibco). Colonies (>50 cells) were counted.
TRANSWELL ASSAYS:
The invasiveness of stably transfected trophoblast cells was evaluated using a Matrigel-coated (BD Biosciences) Transwell system. Briefly, 2×104 trophoblast cells in FBS-free Roswell Park Memorial Institute (RPMI)-1640 medium were seeded into each apical chamber and 200 μL DMEM containing 10% FBS were added to each basolateral chamber. After incubation for 24 h, the non-invasive cells in each apical chamber were removed with a cotton swab. Cells on the lower side of each chamber were fixed in 4% paraformaldehyde (Beyotime), stained with 0.1% crystal violet (Beyotime), and examined with an inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany). Cell numbers were counted in 5 random fields of each chamber and averaged. Migration was assessed using the Transwell system without the Matrigel coating.
CASPASE-3/9 ACTIVITY ASSAY:
Trophoblast cell apoptosis was evaluated using caspase-3 and caspase-9 activity assay kits (Beyotime). Briefly, the transfected trophoblast cell lysates were collected after the addition of lysis buffer (Abgent, Suzhou, Jiangsu, China). Cell lysates were treated with 2 mM Asp-Glu-Val-Asp (to assay caspase-3 activity) or 2 mM Leu-Glu-His-Asp (to assay caspase-9 activity), and labeled with p-nitroaniline for 2 h in the dark at 37°C. The OD value at 405 nm was measured using a Labserv K3 microplate reader (WoYuan, Shanghai, China).
FLOW CYTOMETRY:
Apoptosis was analyzed using Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kits (K201-100, BioVision, Inc., Exton, PA, USA). Trophoblast cells were plated into 6-well plates at a density of 5×104 cells/well for 24 h. The cells were transfected with vector in Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) for 24 h at 37°C, detached with trypsin (Gibco) and resuspended in 1× binding buffer. The cells were doubly stained with Annexin V-FITC and PI for 20 min, and apoptosis was analyzed by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
LUCIFERASE REPORT ASSAY:
PCR-amplified fragments of NEAT1 and the 3′ untranslated region (3′UTR) of FLT1 mRNA that contained predicted miR-373 binding sites were each inserted into the pGL3 luciferase reporter vector (Promega Corporation, Madison, WI, USA) and into the pmirGLO vector (Promega) to produce wild-type (WT)-NEAT1/WT-FLT1 luciferase reporter genes. To generate a corresponding mutant (MT) vector as a control, the seed region of the binding sites with miR-373 were mutated to produce MT-NEAT1/MT-FLT1. Trophoblast cells were co-transfected with the above vectors and miR-373 mimic for 48 h, and luciferase activity was measured with a dual-luciferase assay kit (Promega).
STATISTICAL ANALYSES:
Data were expressed as the mean±standard deviation (SD) of 3 experiments. Differences between 2 groups were compared by unpaired
Results
NEAT1, OVEREXPRESSED IN PLACENTAL TISSUES OF RATS WITH PE, INHIBITS TROPHOBLAST GROWTH:
BP and 24-h urinary protein did not differ in PE and control rats at the beginning of pregnancy (Table 1). At 12 and 18 days, however, both BP and 24-h urinary protein were significantly higher in PE than in control rats.
Immunofluorescence assays of trophoblast cells isolated from placental tissues of rats with PE showed that 94.8% of these cells were positive for cytokeratin 7, whereas 7.8% were positive for vimentin, confirming that these cells were placental trophoblasts (Figure 1A).
RT-qPCR showed that the expression of NEAT1 was significantly higher in placental tissues from PE than from control rats (Figure 1B). To further explore this finding, pcDNA-NEAT1, siRNA-NEAT1, and their controls were transfected into trophoblast cells. Effective transfection was validated by RT-qPCR (Figure 1C), and cell proliferation was measured using CCK-8 and colony formation assays. Transfection of pcDNA-NEAT1 into trophoblasts significantly inhibited cell proliferation, whereas transfection of siRNA-NEAT1 significantly enhanced trophoblast cell proliferation (Figure 1D). Transwell assays showed that pcDNA-NEAT1 markedly reduced, whereas siRNA-NEAT1 markedly enhanced the migration and invasion of trophoblast cells (Figure 1E). Moreover, pcDNA-NEAT1 enhanced the activity of the apoptosis-related genes caspase-3 and caspase-9, whereas siRNA-NEAT1 significantly reduced their activities (Figure 1F), findings consistence with the results of flow cytometric (Figure 1G). Taken together, these findings indicated that NEAT1, which is highly expressed in PE, can inhibit the proliferation, migration and invasion of trophoblast cells, while promote their apoptosis.
MIR-373 IS A TARGET OF NEAT1:
To study the mechanism of NEAT1 action in PE, its subcellular localization was assessed by FISH (Figure 2A) and subcellular fractionation (Figure 2B). Both of these methods showed that NEAT1 is mainly localized in the cytoplasm. Using the bioinformatics website Starbase, we predicted the existence of potential binding sites between miR-373 and NEAT1 (Figure 2C), a finding confirmed by transfecting miR-373 mimic/inhibitor into trophoblast cells, followed by RT-qPCR (Figure 2D). To determine whether NEAT1 bound directly to miR-373, we performed dual-luciferase activity assays by designing WT-NEAT1 and MT-NEAT1 to assess binding capacity. We observed that miR-373 mimic significantly decreased, whereas miR-373 inhibitor significantly increased, the luciferase activity of WT-NEAT1; neither, however, had a significant effect on the luciferase activity of MT-NEAT1 (Figure 2E). These findings suggested the direct binding between NEAT1 and miR-373. Knockdown of NEAT1 expression promoted the expression of miR-373, whereas overexpression of NEAT1 inhibited miR-373 expression (Figure 2F), providing further evidence that NEAT1 binds to miR-373.
MIR-373 IN PLACENTAL TISSUES OF RATS WITH PE PROMOTES GROWTH OF TROPHOBLAST CELLS:
Because RT-qPCR showed that miR-373 expression in placental tissues was much lower in the rats with PE than in normal controls (Figure 3A), we investigated the effects of miR-373 on biological functions in trophoblast cells. As expected, miR-373 knockdown markedly enhanced, whereas miR-373 overexpression markedly inhibited, cell proliferation (Figure 3B). Transwell assays showed that miR-373 mimic facilitated trophoblast cell migration and invasion, both of which were significantly inhibited by miR-373 inhibitor (Figure 3C). Flow cytometry showed that miR-373 mimic markedly inhibited, whereas miR-373 inhibitor significantly enhanced cell apoptosis (Figure 3D). These findings indicate that miR-373 is a direct target of NEAT1 and is involved in the proliferation, invasion, migration, and apoptosis of trophoblast cells.
MIR-373 NEGATIVELY REGULATES THE EXPRESSION OF FLT1:
The bioinformatics website suggested that FLT1 is a potential target of miR-373 (Figure 4A), in agreement with results showing that FLT1 expression enhances the likelihood of PE [14]. The expression of FLT1 was much higher in PE than in control rats (Figure 4B). The ability of miR-373 to bind directly to FLT1 was assessed by dual-luciferase activity assays using WT-FLT1 and MT-FLT1 (Figure 4C). We found that miR-373 mimic significantly reduced, whereas miR-373 inhibitor significantly increased, the luciferase activity of WT-FLT1. In contrast, neither miR-373 mimic nor miR-373 inhibitor had a significant effect on luciferase activity of MT-FLT1. To further explore this finding, miR-373 mimic/inhibitor and their controls were transfected into trophoblast cells, and FLT1 expression was measured by RT-qPCR. miR-373 mimic significantly inhibited, whereas miR-373 inhibitor significantly enhanced, FLT1 expression (Figure 4D), indicating that FLT1 is a target of miR-373.
NEAT1 NEGATIVELY MODULATES MIR-373 BY COMPETITIVELY INTERACTING WITH FLT1 TO INHIBIT TROPHOBLAST CELL GROWTH:
Trophoblast cells were transfected with siRNA-NEAT1, siRNA-NEAT1+miR-373 inhibitor and their respective controls, and FLT1 expression was measured by RT-qPCR (Figure 5A). SiRNA-NEAT1 significantly reduced the expression of FLT1, whereas miR-373 inhibitor reversed this inhibition trend, demonstrating that NEAT1 regulates FLT1 expression by taking up miR-373. Similarly, when trophoblast cells were transfected with pcDNA-NEAT1, pcDNA-NEAT1+siRNA-FLT1 and their respective controls, pcDNA-NEAT1 significantly enhanced FLT1 expression, which was inhibited by siRNA-FLT1 (Figure 5B). CCK-8, colony formation, and Transwell assays showed that the pcDNA-NEAT1-induced inhibition of cell viability, colony formation, invasion, and migration was abrogated by siRNA-FLT1 (Figure 5C, 5D). Moreover, the pro-apoptotic activity of pcDNA-NEAT1 was repressed by siRNA-FLT1 (Figure 5E).
Discussion
Inflammation, oxidative stress, and placental trophoblast cells have all been closely linked to PE. The lack of migration and invasion of trophoblast cells has been found to contribute to inadequate remodeling of spiral arteries, the major driver of PE pathogenesis [15]. Furthermore, lncRNAs have been shown to play pivotal roles in the modulation of trophoblast cell function and the incidence of PE [16,17]. Using a rat model of PE, the present study evaluated the mechanism by which NEAT1 influences the proliferation, migration, invasion, and apoptosis of trophoblast cells through the miR-373/FLT1 axis. Our findings provide forceful evidence that the knockdown of NEAT1 accelerates the proliferation, migration, and invasion of placental trophoblast cells in rats with PE by binding to miR-373 to regulate FLT1.
This study found that miR-373 expression was lower and NEAT1 expression higher in placental tissues and trophoblast cells from rats with PE than in control rats. NEAT1 was observed to be increased in placentas of intrauterine growth-restricted than control pregnancies delivered at term [7], indicating that NEAT1 is associated with pregnancy. Knockdown of NEAT1 was found to inhibit the differentiation of Th17/CD4+ T cell, thus participating in the progression of rheumatoid arthritis, an autoimmune disorder [18]. Nevertheless, the specific role of NEAT1 in PE had not been clearly established. The present study found that knockdown of NEAT1 suppressed apoptosis, as evidenced by lower caspase-3 and caspase-9 activities, while increasing cell viability, migration, and invasion. Similarly, NEAT1 depletion enhanced the viability and reduced apoptosis and caspase-3/9 activities in alveolar epithelial cells challenged with lipopolysaccharide [19]. Moreover, NEAT1 potentiated the angiogenesis and survival of brain microvascular endothelial cells exposed to oxygen-glucose deprivation by targeting miR-377 [20]. However, its association with miR-373 remains largely unclear. miR-373 plays a role in the ceRNA network in ovarian cancers and gliomas [21,22]. Our luciferase reporter assay and RT-qPCR analysis showed that miR-373, which is poorly expressed in PE, is a target of NEAT1 in trophoblast cells. Moreover, the level of miR-373 expression was significantly lower in osteoarthritic than in normal chondrocytes, and upregulation of miR-373 reduced the apoptosis of cultured osteoarthritic chondrocytes [23]. miR-373 expression was also found to be reduced in glioma samples and cells and to be involved in the lncRNA HOXA-AS2-mediated cell viability, migration, and invasion of glioblastoma cells [24]. In addition, we observed that the restoration of miR-373 significantly stimulated trophoblast cell viability, migration, and invasion, mimicking the effects of NEAT1 inhibition in trophoblast cells.
Use of the bioinformatic website Starbase indicated that FLT1 can bind to miR-373, a finding confirmed by RT-qPCR. Interestingly, NEAT1 knockdown can positively regulate FLT1 expression, and miR-373 inhibitor rescued the NEAT1 knockdown-induced reduction in FLT1 expression. FLT1 is 1 of the 2 receptors of vascular endothelial growth factor and is differentially expressed in necrotizing enterocolitis in newborns [25]. In addition, FLT1 expression is enhanced at the end of the first trimester [26]. More recently, miR-145-5p was found to enhance trophoblast cell proliferation and invasion by reducing FLT1 expression [27]. Inhibition of FLT1 by low-dose aspirin may be a promising cellular and molecular mechanism for the prevention of PE [28]. In addition to its effects on trophoblast cells, FLT1 was also shown to be involved in fetoplacental endothelial cell migration and angiogenesis [29], suggesting its tight correlation with pregnancy. Mechanistically, reduced generation of hydrogen sulfide in the placenta was found to facilitate the development of PE by enhancing the release of soluble FLT1 [30]. However, enhanced expression of miR-517a/b and miR-517c in extravillous trophoblast cells during the first trimester led to a marked increase in the release of sFLT1 [31]. Furthermore, our CCK-8, colony formation, and Transwell assays, as well as flow cytometry, provided evidence that the silencing of FLT1 reversed the effects of NEAT1 overexpression on cell proliferation, migration, invasion, and apoptosis, further validating the participation of FLT1 in the ceRNA network.
Conclusions
The present study demonstrated that NEAT1 expression was enhanced in the placental tissues of rats with PE, accelerating the apoptosis and inhibiting the proliferation, invasion, and migration of trophoblast cells. NEAT1 was found to bind to miR-373, preventing it from binding to FLT1 and regulating the activity of the latter. NEAT1 knockdown may be a mediator of trophoblast cell behavior, suggesting NEAT1 as a new potential target for treating PE (Figure 6). Efforts are needed to determine the functional activities of NEAT1
Figures
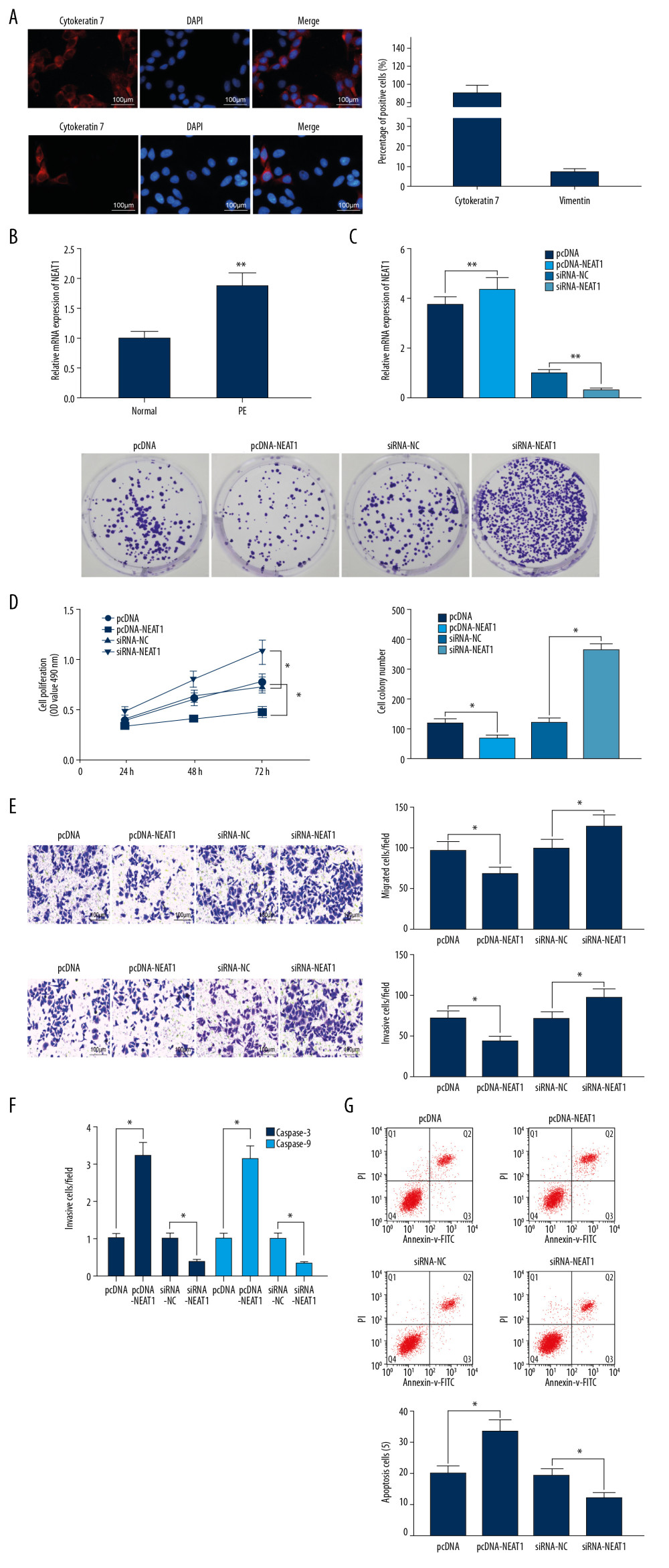 Figure 1. NEAT1 is upregulated in the placenta of PE rats. (A) Immunofluorescence staining, showing the percentages of cells positive for cytokeratin 7 and vimentin proteins; (B) RT-qPCR, showing the expression of NEAT1 mRNA in placental tissues (** p<0.01 by unpaired t test); (C) RT-qPCR, showing the transfection efficiency of pcDNA-NEAT1 and siRNA-NEAT1 (** p<0.01 by 1-way ANOVA); (D) cell proliferation ability, measured by CCK-8 (* p<0.05 by 2-way ANOVA) and colony formation (* p<0.05 by 1-way ANOVA) assays; (E) cell invasion and migration activities measured by Transwell assays (* p<0.05 by 1-way ANOVA); (F) caspase-3 and caspase-9 activities (* p<0.05 by 1-way ANOVA); (G) cell apoptosis, measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 1. NEAT1 is upregulated in the placenta of PE rats. (A) Immunofluorescence staining, showing the percentages of cells positive for cytokeratin 7 and vimentin proteins; (B) RT-qPCR, showing the expression of NEAT1 mRNA in placental tissues (** p<0.01 by unpaired t test); (C) RT-qPCR, showing the transfection efficiency of pcDNA-NEAT1 and siRNA-NEAT1 (** p<0.01 by 1-way ANOVA); (D) cell proliferation ability, measured by CCK-8 (* p<0.05 by 2-way ANOVA) and colony formation (* p<0.05 by 1-way ANOVA) assays; (E) cell invasion and migration activities measured by Transwell assays (* p<0.05 by 1-way ANOVA); (F) caspase-3 and caspase-9 activities (* p<0.05 by 1-way ANOVA); (G) cell apoptosis, measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. 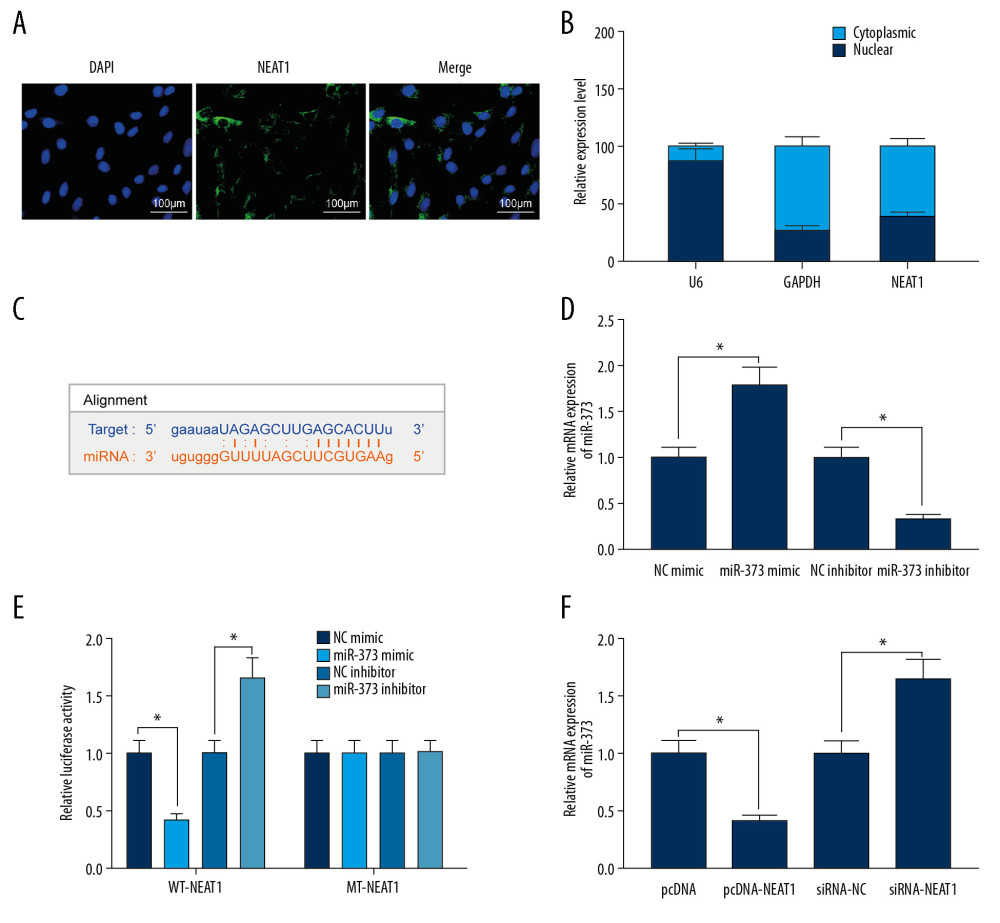 Figure 2. NEAT1 binds to miR-373 and negatively regulates miR-373 expression. (A) FISH, showing that NEAT1 is mainly located in cytoplasm; (B) subcellular fractionation of trophoblast cells, with U6 as a nuclear control and GAPDH as a cytoplasmic control; (C) binding sites of NEAT1 and miR-373; (D) transfection efficiency of miR-373 mimic/inhibitor determined by RT-qPCR (* p<0.05 by unpaired t test); (E) AT1 or MT-NEAT1 (* p<0.05 by 2-way ANOVA); (F) miR-373 expression in cells transfected with pcDNA-NEAT1 or siRNA-NEAT1, as determined by RT-qPCR (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 2. NEAT1 binds to miR-373 and negatively regulates miR-373 expression. (A) FISH, showing that NEAT1 is mainly located in cytoplasm; (B) subcellular fractionation of trophoblast cells, with U6 as a nuclear control and GAPDH as a cytoplasmic control; (C) binding sites of NEAT1 and miR-373; (D) transfection efficiency of miR-373 mimic/inhibitor determined by RT-qPCR (* p<0.05 by unpaired t test); (E) AT1 or MT-NEAT1 (* p<0.05 by 2-way ANOVA); (F) miR-373 expression in cells transfected with pcDNA-NEAT1 or siRNA-NEAT1, as determined by RT-qPCR (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. 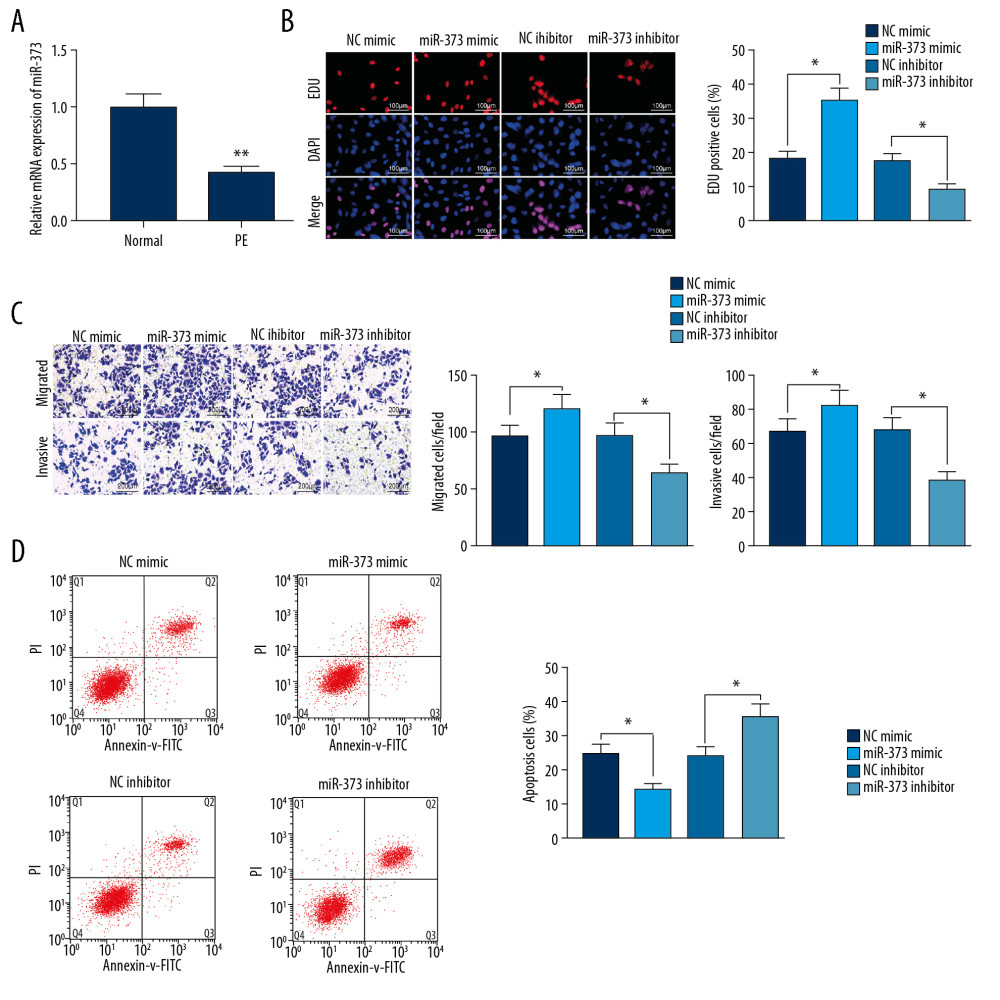 Figure 3. miR-373 enhances trophoblast cell migration and invasion. (A) RT-qPCR measurements of miR-373 expression in placentas of PE and control rats (** p<0.01 by unpaired t test); (B) EdU positivity in trophoblast cells transfected with miR-373 mimic/inhibitor or controls (* p<0.05 by 1-way ANOVA); (C) invasion and migration ability of cells, as measured by Transwell assays (* p<0.05 by 1-way ANOVA); (D) cell apoptosis measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 3. miR-373 enhances trophoblast cell migration and invasion. (A) RT-qPCR measurements of miR-373 expression in placentas of PE and control rats (** p<0.01 by unpaired t test); (B) EdU positivity in trophoblast cells transfected with miR-373 mimic/inhibitor or controls (* p<0.05 by 1-way ANOVA); (C) invasion and migration ability of cells, as measured by Transwell assays (* p<0.05 by 1-way ANOVA); (D) cell apoptosis measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. 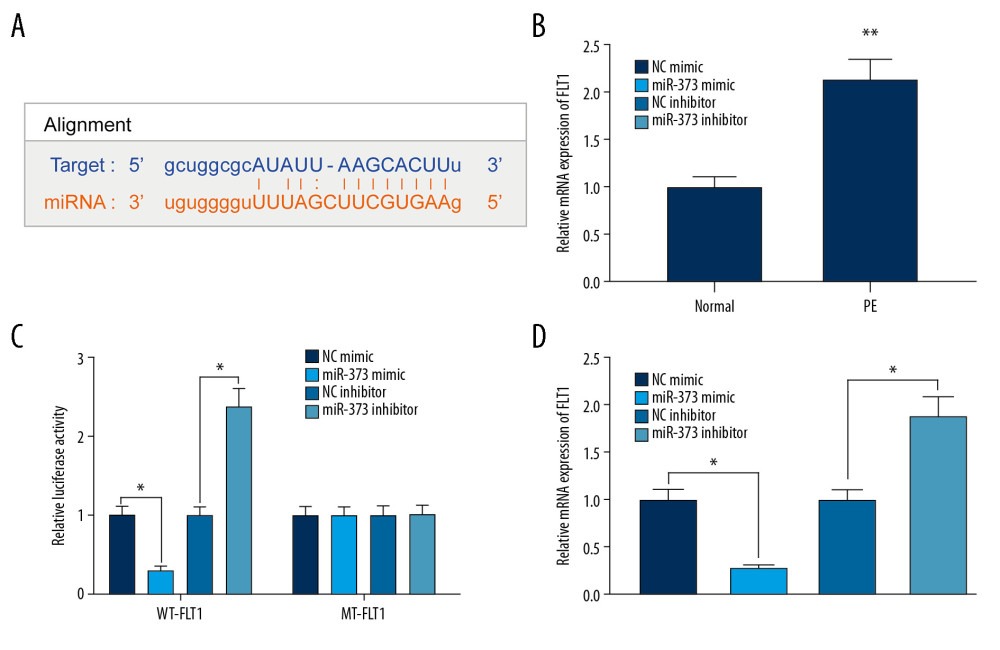 Figure 4. miR-373 directly targets FLT1. (A) Binding sites between miR-373 and FLT1; (B) FLT1 mRNA expression in placentas of PE and control rats evaluated by RT-qPCR (** p<0.01 by unpaired t test); (C) relative luciferase activity in cells co-transfected with miR-373 mimic or miR-373 inhibitor and FLT1-WT-3′UTR or FLT1-MT-3′UTR (* p<0.05 by 2-way ANOVA); (D) FLT1 mRNA expression measured in cells transfected with miR-373 mimic or inhibitor by RT-qPCR (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 4. miR-373 directly targets FLT1. (A) Binding sites between miR-373 and FLT1; (B) FLT1 mRNA expression in placentas of PE and control rats evaluated by RT-qPCR (** p<0.01 by unpaired t test); (C) relative luciferase activity in cells co-transfected with miR-373 mimic or miR-373 inhibitor and FLT1-WT-3′UTR or FLT1-MT-3′UTR (* p<0.05 by 2-way ANOVA); (D) FLT1 mRNA expression measured in cells transfected with miR-373 mimic or inhibitor by RT-qPCR (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. 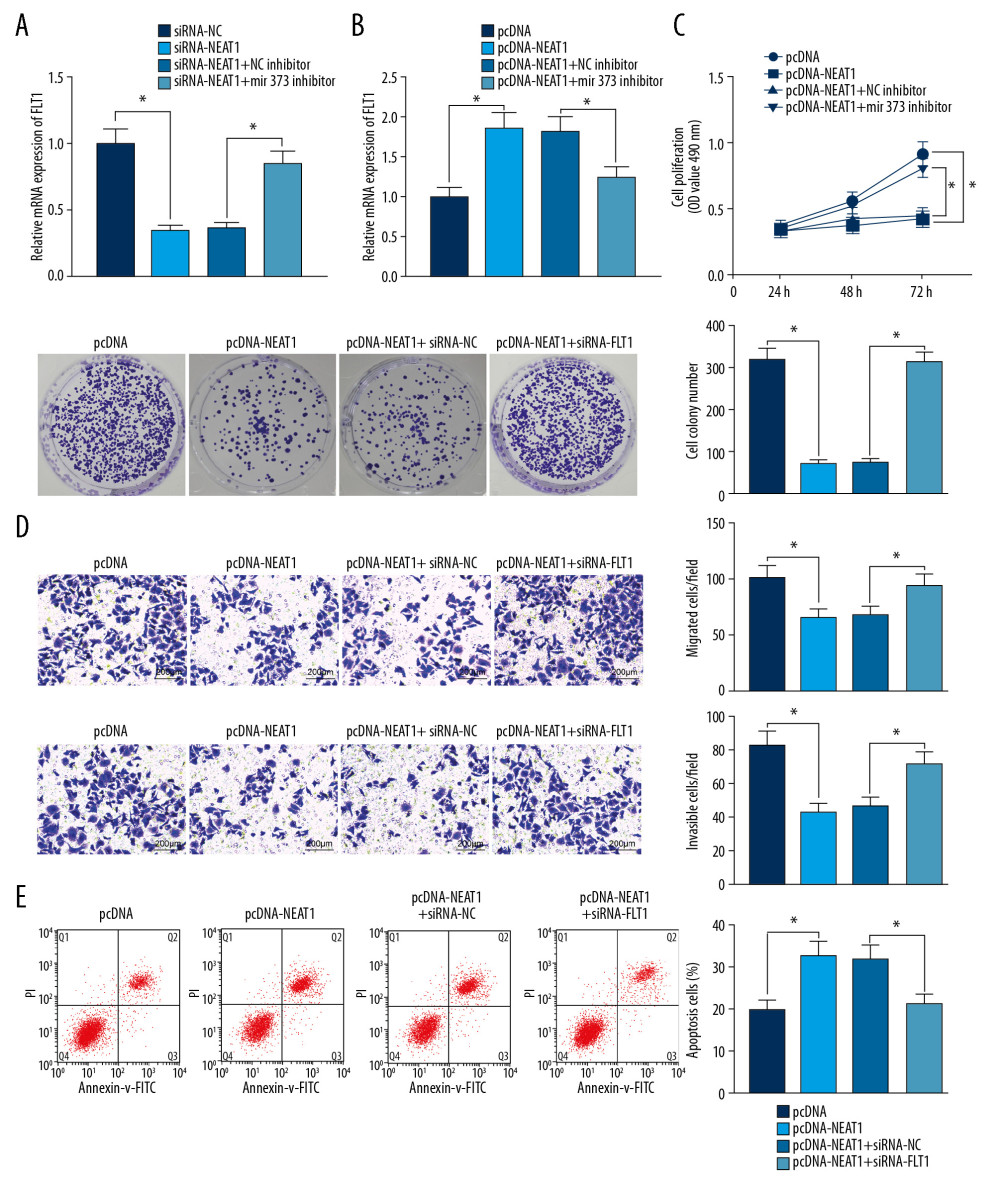 Figure 5. NEAT1 modulates FLT1 via miR-373. (A) RT-qPCR measurements of FLT1 mRNA expression in cells transfected with siRNA-NEAT1 alone or in the presence of miR-373 inhibitor (* p<0.05 by 1-way ANOVA); (B) RT-qPCR measurements of FLT1 mRNA expression in cells transfected with pcDNA-NEAT1 with or without siRNA-FLT1 (* p<0.05 by 1-way ANOVA); (C) cell proliferation activity, measured by the CCK-8 method (* p<0.05 by 2-way ANOVA) and colony formation assay (* p<0.05 by 1-way ANOVA); (D) cell invasion and migration activities, measured by Transwell assays (* p<0.05 by 1-way ANOVA); (E) cell apoptosis, measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 5. NEAT1 modulates FLT1 via miR-373. (A) RT-qPCR measurements of FLT1 mRNA expression in cells transfected with siRNA-NEAT1 alone or in the presence of miR-373 inhibitor (* p<0.05 by 1-way ANOVA); (B) RT-qPCR measurements of FLT1 mRNA expression in cells transfected with pcDNA-NEAT1 with or without siRNA-FLT1 (* p<0.05 by 1-way ANOVA); (C) cell proliferation activity, measured by the CCK-8 method (* p<0.05 by 2-way ANOVA) and colony formation assay (* p<0.05 by 1-way ANOVA); (D) cell invasion and migration activities, measured by Transwell assays (* p<0.05 by 1-way ANOVA); (E) cell apoptosis, measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. 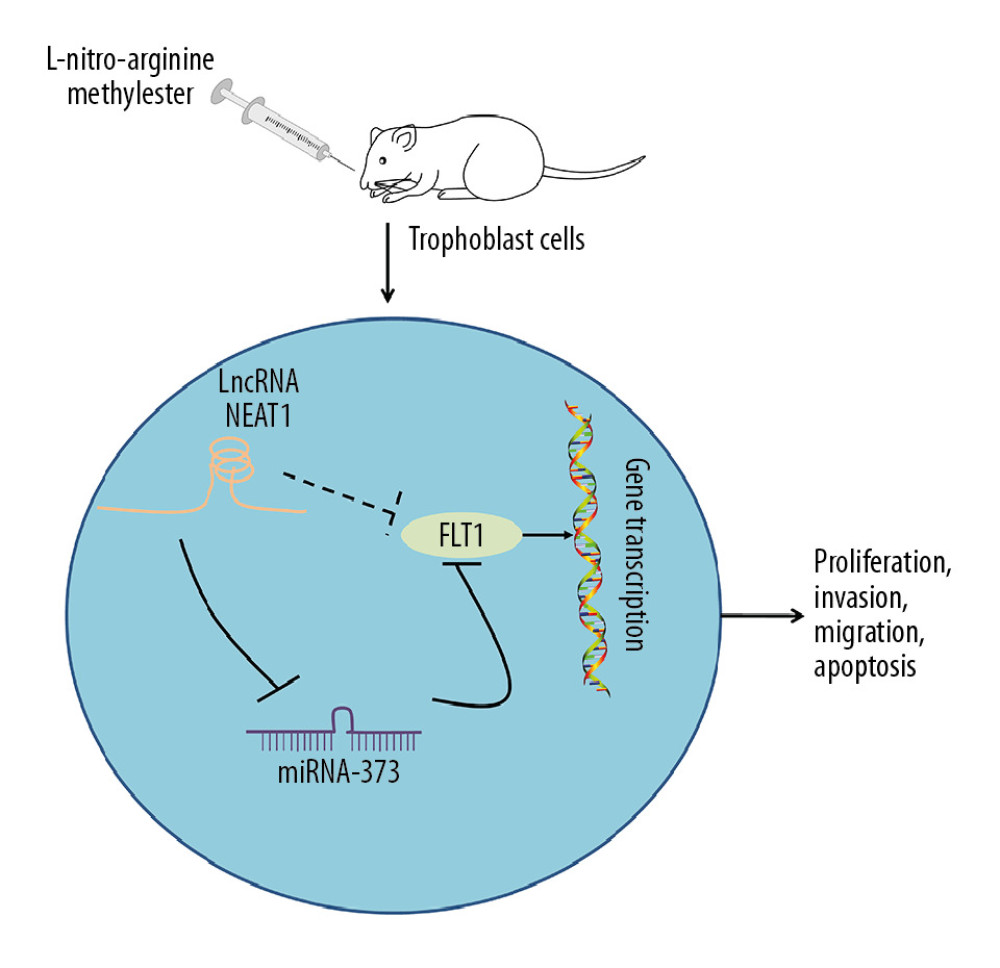 Figure 6. Mechanism of action of the NEAT1/miR-373/FLT1 axis in PE progression. NEAT1 regulates FLT1 expression by binding to miR-373, affecting trophoblast cell proliferation, migration, invasion, and apoptosis.
Figure 6. Mechanism of action of the NEAT1/miR-373/FLT1 axis in PE progression. NEAT1 regulates FLT1 expression by binding to miR-373, affecting trophoblast cell proliferation, migration, invasion, and apoptosis. References
1. El-Sayed AAF, Preeclampsia: A review of the pathogenesis and possible management strategies based on its pathophysiological derangements: Taiwan J Obstet Gynecol, 2017; 56(5); 593-98
2. Phipps E, Prasanna D, Brima W, Jim B, Preeclampsia: Updates in pathogenesis, definitions, and guidelines: Clin J Am Soc Nephrol, 2016; 11(6); 1102-13
3. Travaglino A, Raffone A, Saccone G, Placental morphology, apoptosis, angiogenesis and epithelial mechanisms in early-onset preeclampsia: Eur J Obstet Gynecol Reprod Biol, 2019; 234; 200-6
4. Wang X, Peng S, Cui K, MicroRNA-576-5p enhances the invasion ability of trophoblast cells in preeclampsia by targeting TFAP2A: Mol Genet Genomic Med, 2020; 8(1); e1025
5. Yao RW, Wang Y, Chen LL, Cellular functions of long noncoding RNAs: Nat Cell Biol, 2019; 21(5); 542-51
6. He X, He Y, Xi B, LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis: PLoS One, 2013; 8(11); e81437
7. Gremlich S, Damnon F, Reymondin D, The long non-coding RNA NEAT1 is increased in IUGR placentas, leading to potential new hypotheses of IUGR origin/development: Placenta, 2014; 35(1); 44-49
8. Tay Y, Rinn J, Pandolfi PP, The multilayered complexity of ceRNA crosstalk and competition: Nature, 2014; 505(7483); 344-52
9. Wang WT, Ye H, Wei PP, LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner: J Hematol Oncol, 2016; 9(1); 117
10. Wei F, Cao C, Xu X, Wang J, Diverse functions of miR-373 in cancer: J Transl Med, 2015; 13; 162
11. Sircar M, Thadhani R, Karumanchi SA, Pathogenesis of preeclampsia: Curr Opin Nephrol Hypertens, 2015; 24(2); 131-38
12. Lai WS, Ding YL, GNG7 silencing promotes the proliferation and differentiation of placental cytotrophoblasts in preeclampsia rats through activation of the mTOR signaling pathway: Int J Mol Med, 2019; 43(5); 1939-50
13. Zhao S, Wang J, Cao Z, miR-126a-3p induces proliferation, migration and invasion of trophoblast cells in pre-eclampsia-like rats by inhibiting a disintegrin and metalloprotease 9: Biosci Rep, 2019; 39(12); BSR20191271
14. Austdal M, Silva GB, Bowe S, Metabolomics identifies placental dysfunction and confirms Flt-1 (FMS-like tyrosine kinase receptor 1) biomarker specificity: Hypertension, 2019; 74(5); 1136-43
15. Gong F, Cheng H, Shi Y, LncRNA TDRG1/miR-214-5p axis affects preeclampsia by modulating trophoblast cells: Cell Biochem Funct, 2020; 38(4); 352-61
16. Zhang Y, Zou Y, Wang W, Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells: J Cell Biochem, 2015; 116(4); 542-50
17. Zuo Q, Huang S, Zou Y, The Lnc RNA SPRY4-IT1 modulates trophoblast cell invasion and migration by affecting the epithelial-mesenchymal transition: Sci Rep, 2016; 6; 37183
18. Shui X, Chen S, Lin J, Knockdown of lncRNA NEAT1 inhibits Th17/CD4(+) T cell differentiation through reducing the STAT3 protein level: J Cell Physiol, 2019; 234(12); 22477-84
19. Zhou H, Wang X, Zhang B, Depression of lncRNA NEAT1 antagonizes LPS-evoked acute injury and inflammatory response in alveolar epithelial cells via HMGB1-RAGE signaling: Mediators Inflamm, 2020; 2020; 8019467
20. Zhou ZW, Zheng LJ, Ren X, LncRNA NEAT1 facilitates survival and angiogenesis in oxygen-glucose deprivation (OGD)-induced brain microvascular endothelial cells (BMECs) via targeting miR-377 and upregulating SIRT1, VEGFA, and BCL-XL: Brain Res, 2019; 1707; 90-98
21. Zhang Z, Cheng J, Wu Y, LncRNA HOTAIR controls the expression of Rab22a by sponging miR-373 in ovarian cancer: Mol Med Rep, 2016; 14(3); 2465-72
22. Zhou XY, Liu H, Ding ZB, lncRNA SNHG16 promotes glioma tumorigenicity through miR-373/EGFR axis by activating PI3K/AKT pathway: Genomics, 2020; 112(1); 1021-29
23. Song J, Kim D, Chun CH, Jin EJ, miR-370 and miR-373 regulate the pathogenesis of osteoarthritis by modulating one-carbon metabolism via SHMT-2 and MECP-2, respectively: Aging Cell, 2015; 14(5); 826-37
24. Gao Y, Yu H, Liu Y, Long non-coding RNA HOXA-AS2 regulates malignant glioma behaviors and vasculogenic mimicry formation via the miR-373/EGFR axis: Cell Physiol Biochem, 2018; 45(1); 131-47
25. Liu H, Wang YB, Systematic large-scale meta-analysis identifies miRNA-429/200a/b and miRNA-141/200c clusters as biomarkers for necrotizing enterocolitis in newborn: Biosci Rep, 2019; 39(9); BSR20191503
26. Ohkuchi A, Hirashima C, Suzuki H, Evaluation of a new and automated electrochemiluminescence immunoassay for plasma sFlt-1 and PlGF levels in women with preeclampsia: Hypertens Res, 2010; 33(5); 422-27
27. Lv Y, Lu X, Li C, miR-145-5p promotes trophoblast cell growth and invasion by targeting FLT1: Life Sci, 2019; 239; 117008
28. Lin L, Li G, Zhang W, Low-dose aspirin reduces hypoxia-induced sFlt1 release via the JNK/AP-1 pathway in human trophoblast and endothelial cells: J Cell Physiol, 2019; 234(10); 18928-41
29. Ji S, Xin H, Li Y, Su EJ, FMS-like tyrosine kinase 1 (FLT1) is a key regulator of fetoplacental endothelial cell migration and angiogenesis: Placenta, 2018; 70; 7-14
30. Hu TX, Guo X, Wang G, MiR133b is involved in endogenous hydrogen sulfide suppression of sFlt-1 production in human placenta: Placenta, 2017; 52; 33-40
31. Anton L, Olarerin-George AO, Hogenesch JB, Elovitz MA, Placental expression of miR-517a/b and miR-517c contributes to trophoblast dysfunction and preeclampsia: PLoS One, 2015; 10(3); e0122707
Figures
 Figure 1. NEAT1 is upregulated in the placenta of PE rats. (A) Immunofluorescence staining, showing the percentages of cells positive for cytokeratin 7 and vimentin proteins; (B) RT-qPCR, showing the expression of NEAT1 mRNA in placental tissues (** p<0.01 by unpaired t test); (C) RT-qPCR, showing the transfection efficiency of pcDNA-NEAT1 and siRNA-NEAT1 (** p<0.01 by 1-way ANOVA); (D) cell proliferation ability, measured by CCK-8 (* p<0.05 by 2-way ANOVA) and colony formation (* p<0.05 by 1-way ANOVA) assays; (E) cell invasion and migration activities measured by Transwell assays (* p<0.05 by 1-way ANOVA); (F) caspase-3 and caspase-9 activities (* p<0.05 by 1-way ANOVA); (G) cell apoptosis, measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 1. NEAT1 is upregulated in the placenta of PE rats. (A) Immunofluorescence staining, showing the percentages of cells positive for cytokeratin 7 and vimentin proteins; (B) RT-qPCR, showing the expression of NEAT1 mRNA in placental tissues (** p<0.01 by unpaired t test); (C) RT-qPCR, showing the transfection efficiency of pcDNA-NEAT1 and siRNA-NEAT1 (** p<0.01 by 1-way ANOVA); (D) cell proliferation ability, measured by CCK-8 (* p<0.05 by 2-way ANOVA) and colony formation (* p<0.05 by 1-way ANOVA) assays; (E) cell invasion and migration activities measured by Transwell assays (* p<0.05 by 1-way ANOVA); (F) caspase-3 and caspase-9 activities (* p<0.05 by 1-way ANOVA); (G) cell apoptosis, measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. Figure 2. NEAT1 binds to miR-373 and negatively regulates miR-373 expression. (A) FISH, showing that NEAT1 is mainly located in cytoplasm; (B) subcellular fractionation of trophoblast cells, with U6 as a nuclear control and GAPDH as a cytoplasmic control; (C) binding sites of NEAT1 and miR-373; (D) transfection efficiency of miR-373 mimic/inhibitor determined by RT-qPCR (* p<0.05 by unpaired t test); (E) AT1 or MT-NEAT1 (* p<0.05 by 2-way ANOVA); (F) miR-373 expression in cells transfected with pcDNA-NEAT1 or siRNA-NEAT1, as determined by RT-qPCR (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 2. NEAT1 binds to miR-373 and negatively regulates miR-373 expression. (A) FISH, showing that NEAT1 is mainly located in cytoplasm; (B) subcellular fractionation of trophoblast cells, with U6 as a nuclear control and GAPDH as a cytoplasmic control; (C) binding sites of NEAT1 and miR-373; (D) transfection efficiency of miR-373 mimic/inhibitor determined by RT-qPCR (* p<0.05 by unpaired t test); (E) AT1 or MT-NEAT1 (* p<0.05 by 2-way ANOVA); (F) miR-373 expression in cells transfected with pcDNA-NEAT1 or siRNA-NEAT1, as determined by RT-qPCR (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. Figure 3. miR-373 enhances trophoblast cell migration and invasion. (A) RT-qPCR measurements of miR-373 expression in placentas of PE and control rats (** p<0.01 by unpaired t test); (B) EdU positivity in trophoblast cells transfected with miR-373 mimic/inhibitor or controls (* p<0.05 by 1-way ANOVA); (C) invasion and migration ability of cells, as measured by Transwell assays (* p<0.05 by 1-way ANOVA); (D) cell apoptosis measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 3. miR-373 enhances trophoblast cell migration and invasion. (A) RT-qPCR measurements of miR-373 expression in placentas of PE and control rats (** p<0.01 by unpaired t test); (B) EdU positivity in trophoblast cells transfected with miR-373 mimic/inhibitor or controls (* p<0.05 by 1-way ANOVA); (C) invasion and migration ability of cells, as measured by Transwell assays (* p<0.05 by 1-way ANOVA); (D) cell apoptosis measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. Figure 4. miR-373 directly targets FLT1. (A) Binding sites between miR-373 and FLT1; (B) FLT1 mRNA expression in placentas of PE and control rats evaluated by RT-qPCR (** p<0.01 by unpaired t test); (C) relative luciferase activity in cells co-transfected with miR-373 mimic or miR-373 inhibitor and FLT1-WT-3′UTR or FLT1-MT-3′UTR (* p<0.05 by 2-way ANOVA); (D) FLT1 mRNA expression measured in cells transfected with miR-373 mimic or inhibitor by RT-qPCR (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 4. miR-373 directly targets FLT1. (A) Binding sites between miR-373 and FLT1; (B) FLT1 mRNA expression in placentas of PE and control rats evaluated by RT-qPCR (** p<0.01 by unpaired t test); (C) relative luciferase activity in cells co-transfected with miR-373 mimic or miR-373 inhibitor and FLT1-WT-3′UTR or FLT1-MT-3′UTR (* p<0.05 by 2-way ANOVA); (D) FLT1 mRNA expression measured in cells transfected with miR-373 mimic or inhibitor by RT-qPCR (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. Figure 5. NEAT1 modulates FLT1 via miR-373. (A) RT-qPCR measurements of FLT1 mRNA expression in cells transfected with siRNA-NEAT1 alone or in the presence of miR-373 inhibitor (* p<0.05 by 1-way ANOVA); (B) RT-qPCR measurements of FLT1 mRNA expression in cells transfected with pcDNA-NEAT1 with or without siRNA-FLT1 (* p<0.05 by 1-way ANOVA); (C) cell proliferation activity, measured by the CCK-8 method (* p<0.05 by 2-way ANOVA) and colony formation assay (* p<0.05 by 1-way ANOVA); (D) cell invasion and migration activities, measured by Transwell assays (* p<0.05 by 1-way ANOVA); (E) cell apoptosis, measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates.
Figure 5. NEAT1 modulates FLT1 via miR-373. (A) RT-qPCR measurements of FLT1 mRNA expression in cells transfected with siRNA-NEAT1 alone or in the presence of miR-373 inhibitor (* p<0.05 by 1-way ANOVA); (B) RT-qPCR measurements of FLT1 mRNA expression in cells transfected with pcDNA-NEAT1 with or without siRNA-FLT1 (* p<0.05 by 1-way ANOVA); (C) cell proliferation activity, measured by the CCK-8 method (* p<0.05 by 2-way ANOVA) and colony formation assay (* p<0.05 by 1-way ANOVA); (D) cell invasion and migration activities, measured by Transwell assays (* p<0.05 by 1-way ANOVA); (E) cell apoptosis, measured by flow cytometry (* p<0.05 by 1-way ANOVA). Each figure represents the average of 3 replicates. Figure 6. Mechanism of action of the NEAT1/miR-373/FLT1 axis in PE progression. NEAT1 regulates FLT1 expression by binding to miR-373, affecting trophoblast cell proliferation, migration, invasion, and apoptosis.
Figure 6. Mechanism of action of the NEAT1/miR-373/FLT1 axis in PE progression. NEAT1 regulates FLT1 expression by binding to miR-373, affecting trophoblast cell proliferation, migration, invasion, and apoptosis. In Press
05 Mar 2024 : Clinical Research
Role of Critical Shoulder Angle in Degenerative Type Rotator Cuff Tears: A Turkish Cohort StudyMed Sci Monit In Press; DOI: 10.12659/MSM.943703
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952