24 October 2021: Clinical Research
Vitamin D Receptor Is a Sepsis-Susceptibility Gene in Chinese Children
Danni He12BCDEF, Xiuxiu Lu3ABCDEFG, Wei Li3BG, Yuanyuan Wang4BD, Ning Li3BC, Yuanmei Chen2CEG, Lipeng Zhang25EF, Wenquan Niu1AEF*, Qi Zhang2AEGDOI: 10.12659/MSM.932518
Med Sci Monit 2021; 27:e932518
Abstract
BACKGROUND: We designed an association study among 267 cases of children with sepsis and 283 healthy controls, by genotyping 9 variants in the VDR gene.
MATERIAL AND METHODS: This was a hospital-based, case-control, genetic association study. In addition to 3 genetic modes of inheritance, haplotype and interaction analyses were employed to examine the prediction of VDR gene for pediatric sepsis. Effect-size estimates are expressed as odds ratio (OR) and 95% confidence interval (CI).
RESULTS: Two variants in the VDR gene, rs2107301 and rs2189480, were found to play a leading role in susceptibility to sepsis in children. The mutant homozygotes of rs2107301 (CC) and rs2189480 (CC) were associated with a reduced risk of sepsis compared with the corresponding wild homozygotes (OR: 0.44 and 0.43, 95% CI: 0.21-0.92 and 0.23-0.81, p: 0.03 and 0.009, respectively). The mutations of rs2107301-C and rs2189480-C alleles were associated with reduced sepsis risk. Haplotype C-C-C-C-C-T-C-A-G in the VDR gene was significantly associated with a 0.59-fold decreased risk of sepsis (95% CI: 0.12-0.76, p: 0.02). In the haplotype–phenotype analysis, significant association was noted for high-density lipoprotein, even after simulation correction (psim <0.05).
CONCLUSIONS: Taken together, our findings indicate that the VDR gene may be a sepsis-susceptibility gene in Chinese Han children.
Keywords: Adult Children, MED4 Protein, Human, Risk Assessment, Sepsis, Case-Control Studies, Child, Child, Preschool, Genetic Association Studies, Genetic Predisposition to Disease, Humans, Infant, Polymorphism, Single Nucleotide, Receptors, Calcitriol
Background
Sepsis is commonly seen among critically ill children worldwide, with a prevalence rate of 8.2% and an in-hospital mortality rate as high as 25% [1]. The Resolution on Sepsis by the United Nations World Health Assembly in 2017 recognized sepsis as a global threat in children and a priority to address during the next decade [2]. Sepsis is a polygenic and multifactor symptom, with ambiguous etiology [3]. There is evidence from studies of twins with late-onset sepsis [4,5] showing that genetic variability may influence the susceptibility to sepsis through the innate immune system. These genetic variants explain different outcomes of pediatric patients under standardized treatments, and also provide important clues for new mechanisms of sepsis.
The candidate gene approach assumes that the gene with a known biological function is the host one regulating the investigated traits [6,7]. Using this approach, our previous study revealed that the gene encoding vitamin D receptor (VDR) may be a candidate for sepsis risk, as some variants showed a cumulative effect on neonatal sepsis cases [8]. VDR is a kind of nuclear receptor that plays a central role in 1α, 25-dihydroxyvitamin D3’s biological actions, and regulates mass gene expression, cellular proliferation and differentiation, and immune response, largely in a ligand-dependent manner [9,10].
Animal experiments have shown that the activation of VDR can protect or attenuate organ injury through inhibiting cell apoptosis, and has even been shown in a mouse model to reverse sepsis-induced immunosuppression through enhancing autophagy [11–13]. Therefore, we developed the hypothesis that the
Material and Methods
STUDY CHILDREN:
This study was a hospital-based, case-control, genetic association study. Recruitment was carried out at the Emergency Department and Intensive Care Unit (ICU) of the Capital Institute of Pediatrics, Beijing, China. A total of 267 children with sepsis who met the criteria for treatment during the period from October 2017 to April 2020 and 283 healthy controls were included. The conduct of this study was approved by the Ethics Review Committee of the Capital Institute of Pediatrics in Beijing, China (approval ID SHERLL 2013075). All participants read and signed the informed consent form. If children were unable to sign the consent form, the guardians signed on their behalf. This study complied with the Declaration of Helsinki.
INCLUSION AND EXCLUSION CRITERIA:
The inclusion criteria were: a) patients under 12 years old admitted to the Pediatric Intensive Care Unit (PICU) diagnosed with sepsis; b) patients were included if they fulfilled criteria of the International Pediatric Sepsis Consensus Conference: Definitions for sepsis and organ dysfunction in pediatrics [14]. The exclusion criteria were as follows: a) patients with known autoimmune disease and cancer; b) using immunosuppressant or immunomodulator; c) congenital organ dysfunction and d) diagnosis of sepsis or shock over 72 h.
DNA EXTRACTION AND QUALITY CONTROL:
Venous blood samples were taken in 5-mL Vacutainer tubes with K3-EDTA from each participant. Plasma was separated by centrifugation at 4°C, then frozen in a freezer at −80°C. The RelaxGene Blood DNA System (Tiangen Biotech, Beijing, China) was used to extract genomic DNA from white blood cells according to the manufacturers’ guidelines. The specific process of DNA extraction is provided in Supplementary File 1. Then, we used a spectrophotometer to determine concentration (at A260 nm) and purity (at A260/A280 ratio) of DNA.
VARIANT SELECTION:
Nine variants in the VDR gene were selected: rs9729, rs2107301, rs2189480, rs2239185, rs3782905, rs4516035, rs7139166, rs11168266, and rs11168293. The selection of these variants was based on published papers [15–19] and the NCBI-Gene website analysis (https://www.ncbi.nlm.nih.gov/gene/).
GENOTYPING:
The 9 variants in the VDR gene were amplified by polymerase chain reaction (PCR) and sequenced by 3730 sequencing analysis. The primer sequences were designed according to the genomic sequence deposited in the NCBI database; 10% of the samples were randomly selected and re-genotyped to ensure the consistency of results. Sequencing results’ alignment and multiple comparisons were analyzed by Chromas Lite version 2.01 (http://www.technelysium.com.au). PCR was performed using the following parameters: denaturation for 5 min at 95°C, 30 cycles of 95°C for 15 s, Tm°C for 15 s (annealing temperatures are provided in Supplementary File 2), and extension for 1 min at 72°C, with a final extension for 7 min at 72°C. The DNA extraction procedure and primer sequences are provided in Supplementary File 2.
STATISTICAL ANALYSIS:
For database management and statistical analysis, we used STATA software Release 14.1 (Stata Corp, TX). Continuous variables are expressed as mean (standard deviation), and compared using the
Generally, a haplotype is a combination of multiple alleles on one chromosome. We did some haplotype-based statistical analysis to explore the interactions of these variants. The Haplo.em program was used to compute the haplotype frequencies for the variants in different groups. Haplo.glm and haplo.cc were used to calculate effect sizes for each variant and haplo.score was used to estimate an individual’s phenotype as a function of each inferred haplotype. Simulate P (
Interaction analysis was implemented using the open-source multifactor dimensionality reduction (MDR) software package Release 3.0.2 available from
Results
BASELINE CHARACTERISTICS:
Table 1 shows the baseline characteristics of sepsis cases and healthy controls. Controls were significantly older than cases (45 months vs 29 months, p<0.001), and males were overrepresented among cases.
SINGLE VARIANT ANALYSIS:
Table 2 shows the genotype distributions and allele frequencies of 9 variants in the VDR gene. The genotype distributions differed very significantly for rs2107301 and rs2189480 between cases and controls (p: 0.01 and 0.004, separately). The mutant allele frequencies of rs2107301 (C) and rs2189480 (C) were significantly higher in healthy controls than in sepsis groups (p: 0.003 and 0.001, respectively). There was no hint of significant differences in the other 7 variations, either in genotype distributions or allele frequencies, between the 2 groups. The effect size of each variations’ genotype was calculated using their wild homozygous genotype as a reference (Table 2). The mutant homozygous genotypes of rs2107301 (CC) and rs2189480 (CC) were associated with reduced risk compared to the wild homozygous genotype (OR: 0.44 and 0.43, 95% CI: 0.21–0.92 and 0.23–0.81, p: 0.03 and 0.009, respectively). Among the genotypes of rs2189480, the CA genotype also showed a lower risk of sepsis than the AA type (OR: 0.62, 95% CI: 0.43–0.90, p: 0.01). In the genotypes of the remaining variations, the differences in effect size were not significant. All p values were calculated after adjusting for age and sex in logistic regression analysis.
The unadjusted and adjusted risk predictions of the 9 studied variants in the VDR gene for sepsis mortality risk were calculated in both additive and dominant models to correct for some mutant homozygotes that were not sufficiently numerous, which has an impact on the predictive value (Table 3). In general, 2 variants, rs2107301 and rs2189480, showed significant protective effects under the additive and dominant model. The effect sizes were: rs2107301 in the additive model (OR: 0.68, 95% CI: 0.51–0.91, p: 0.008), rs2107301 in the dominant model (OR: 0.65, 95% CI: 0.46–0.93, p: 0.02), rs2189480 in the additive model (OR: 0.64, 95% CI: 0.49–0.84, p: 0.001), and rs2189480 in the dominant model (OR: 0.58, 95% CI: 0.41–0.83, p: 0.003). All the results were meaningful regardless of whether factors were adjusted or not.
HAPLOTYPE ANALYSIS AND HAPLOTYPE–PHENOTYPE ASSOCIATION:
Table 4 presents the derived haplotype frequencies and risk estimates for pediatric sepsis. Haplotype C-T-A-C-C-T-C-G-G (alleles arranged by order of rs9729, rs2107301, rs2189480, rs2239185, rs3782905, rs4516035, rs7139166, rs11168266, and rs11168293, with the same hereafter) was the most common (frequency less than 1% is not displayed). Frequency of haplotype C-C-C-C-C-T-C-A-G was significantly higher in controls than in cases (4% vs 1%, p: 0.02) and was significantly associated with a 0.59-fold decreased risk of pediatric sepsis (95% CI: 0.12–0.76, p: 0.02). Other haplotypes had no significant association with sepsis risk.
We explored a haplotype–phenotype association by taking all haplotypes of the 9 studied variants as a whole, and tested the comprehensive correlation of haplotypes with all collected baseline characteristics (Table 5). A significant association was noted for high-density lipoprotein (HDL) in sepsis patients after simulation correction (psim<0.05).
INTERACTION ANALYSIS:
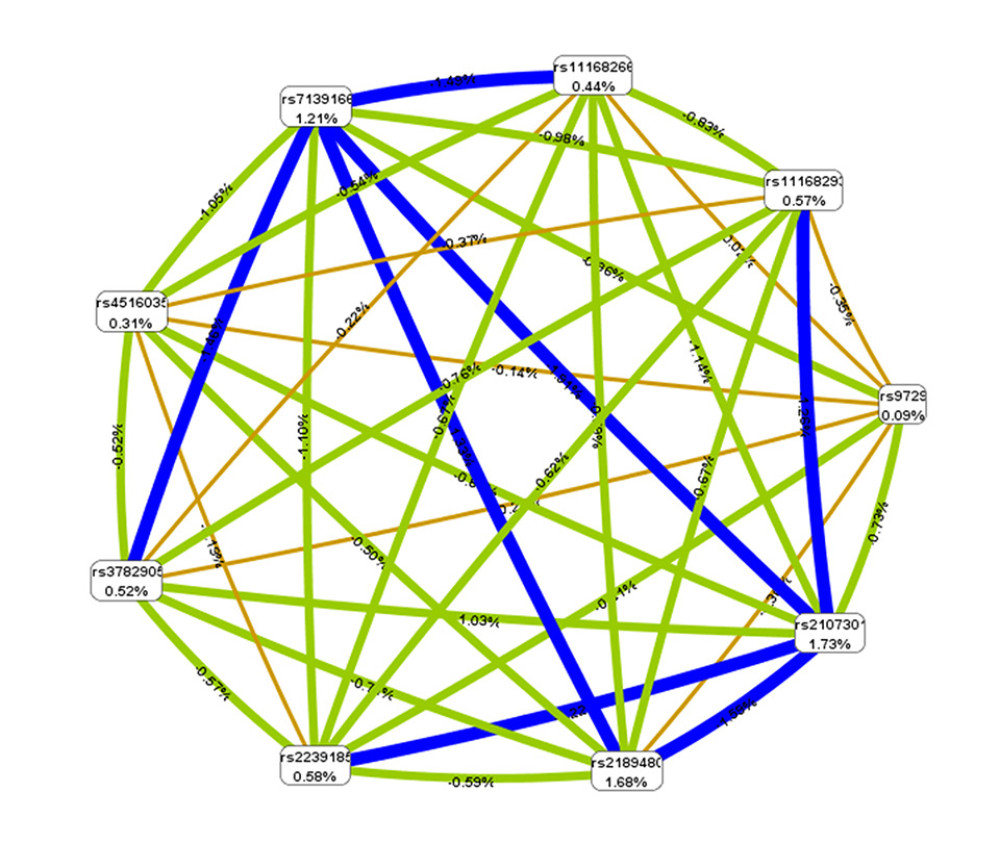
The interaction of the 9 variants under study in predisposition to sepsis in children is displayed in Figure 1. Blue lines and green lines represent antagonism, and the intensity of blue was stronger than that of green; red lines or orange lines represent synergistic effect, and the intensity of red was stronger than that of orange (this study did not produce red or orange lines, indicating that antagonism was dominant among various sites). Overall, there was no evidence of synergistic interaction between variants.
Discussion
LIMITATIONS:
Several limitations should be acknowledged for this study. Firstly, there was a significant difference between groups in sex and age. Although we carried out statistical adjustments, the bias cannot be eliminated. Secondly, as an observational case-control association study, our results cannot prove the cause-effect relationship between
Conclusions
Taken together, our findings indicate that the
Tables
Table 1. The baseline characteristics of study children.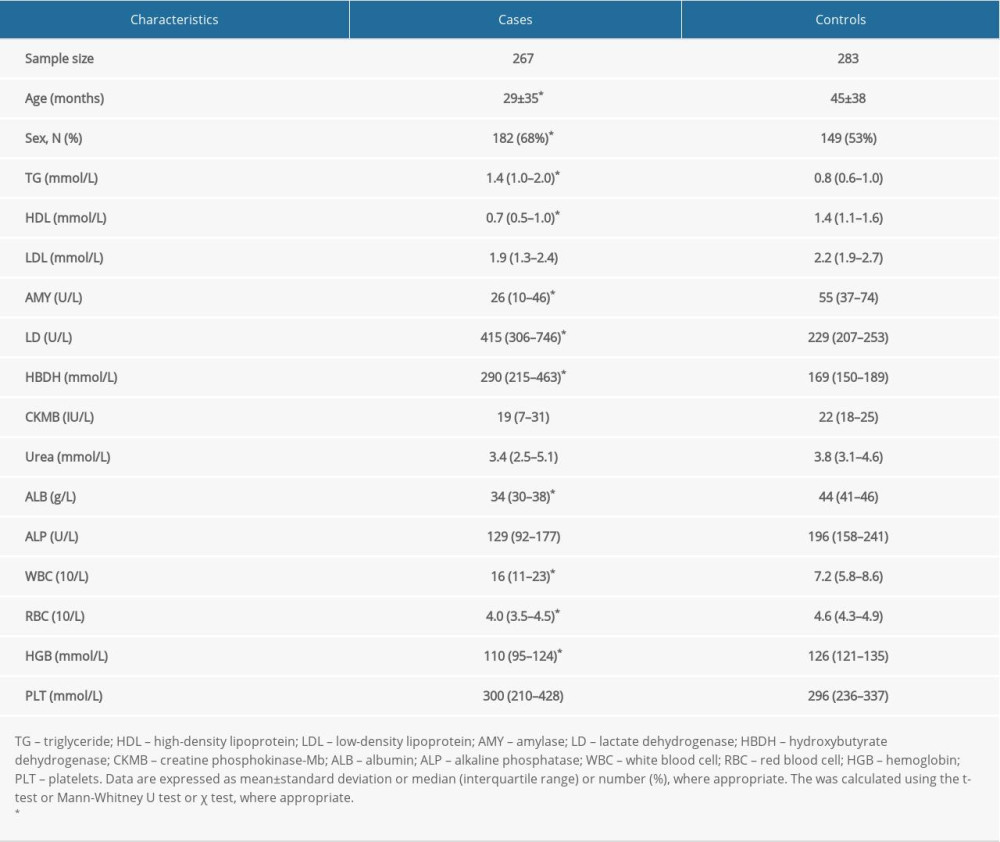 Table 2. Genotype and allele distributions of VDR gene 9 studied variants between cases and controls, and genotype-based risk prediction for sepsis mortality risk.
Table 2. Genotype and allele distributions of VDR gene 9 studied variants between cases and controls, and genotype-based risk prediction for sepsis mortality risk.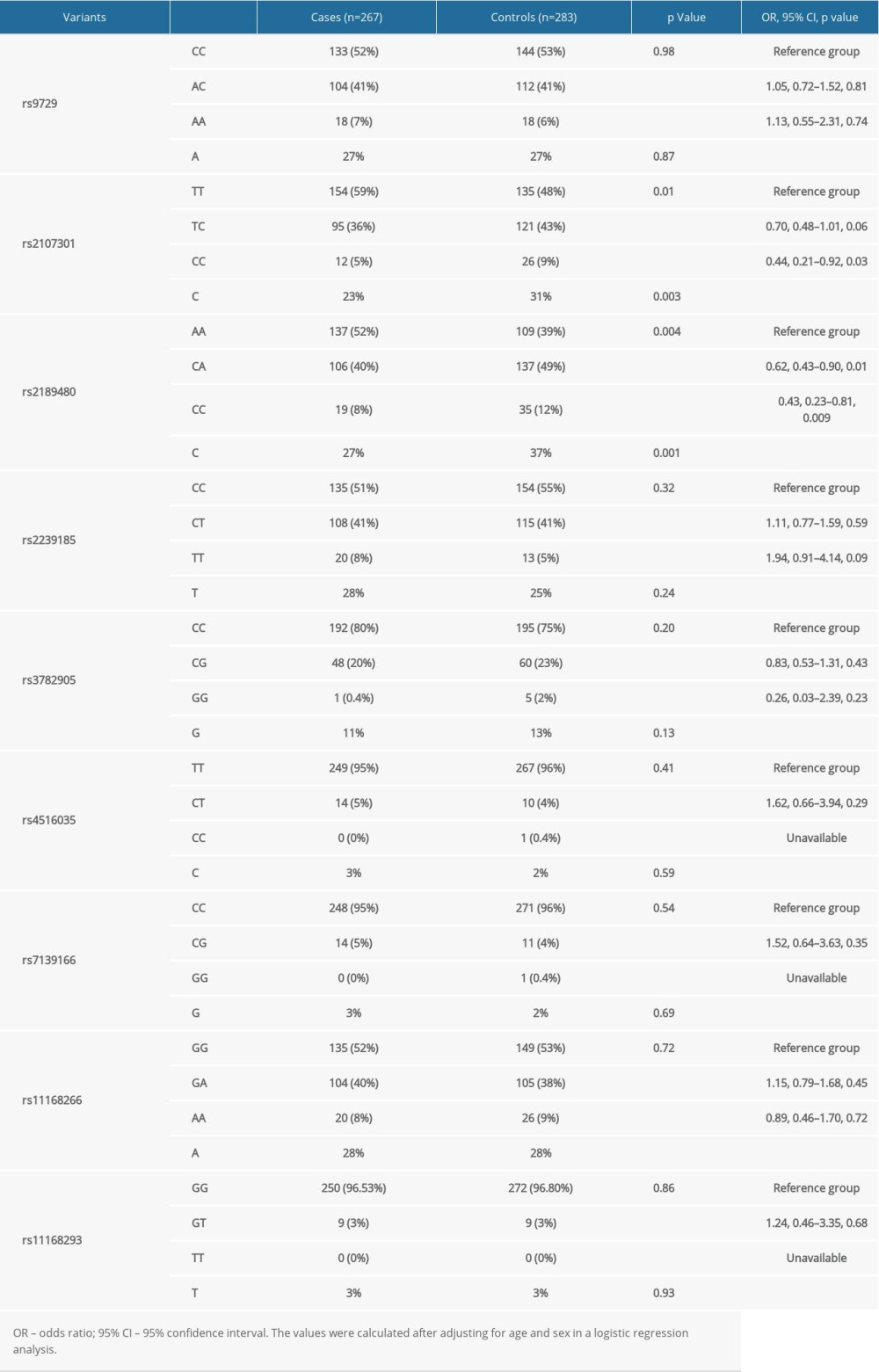 Table 3. The unadjusted and adjusted risk prediction of 9 studied variants for sepsis mortality risk under additive and dominant models, respectively.
Table 3. The unadjusted and adjusted risk prediction of 9 studied variants for sepsis mortality risk under additive and dominant models, respectively.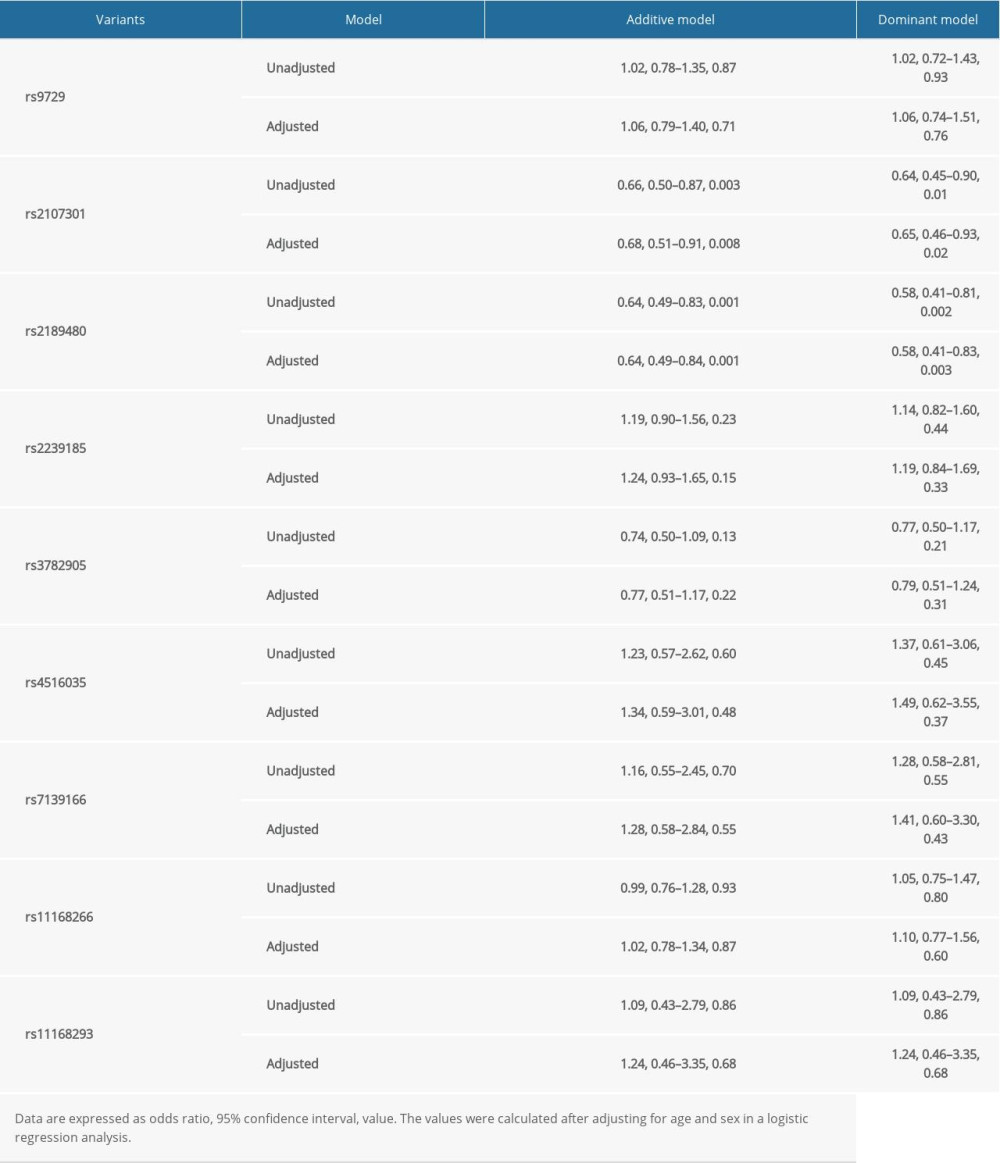 Table 4. Haplotype frequencies (>1% in all cases and controls) of variants in VDR genes between cases and controls, and haplotype-based risk prediction for sepsis mortality risk.
Table 4. Haplotype frequencies (>1% in all cases and controls) of variants in VDR genes between cases and controls, and haplotype-based risk prediction for sepsis mortality risk.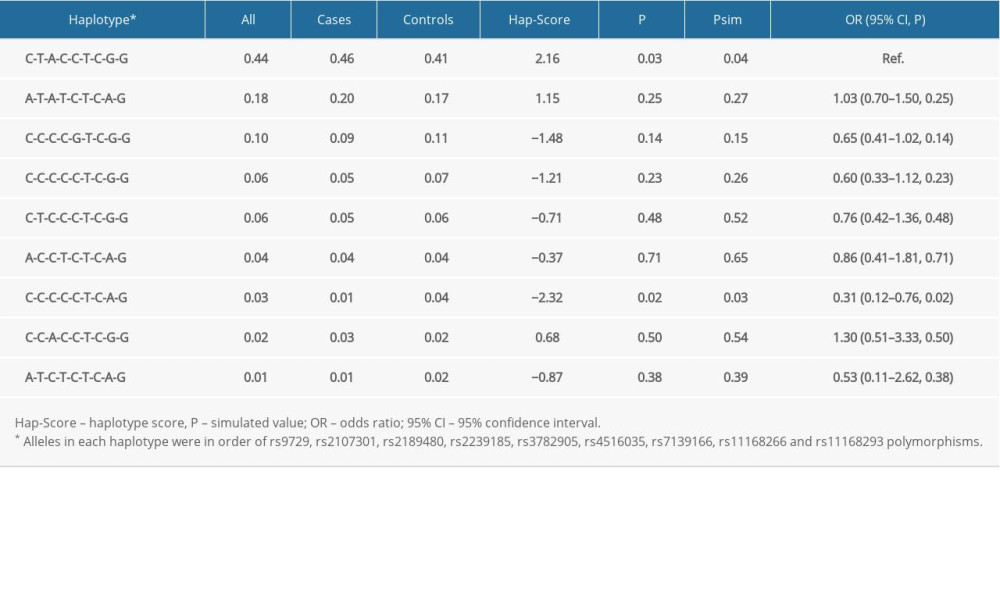 Table 5. Global testing of all haplotypes with anthropometric index and clinical biomarkers.
Table 5. Global testing of all haplotypes with anthropometric index and clinical biomarkers.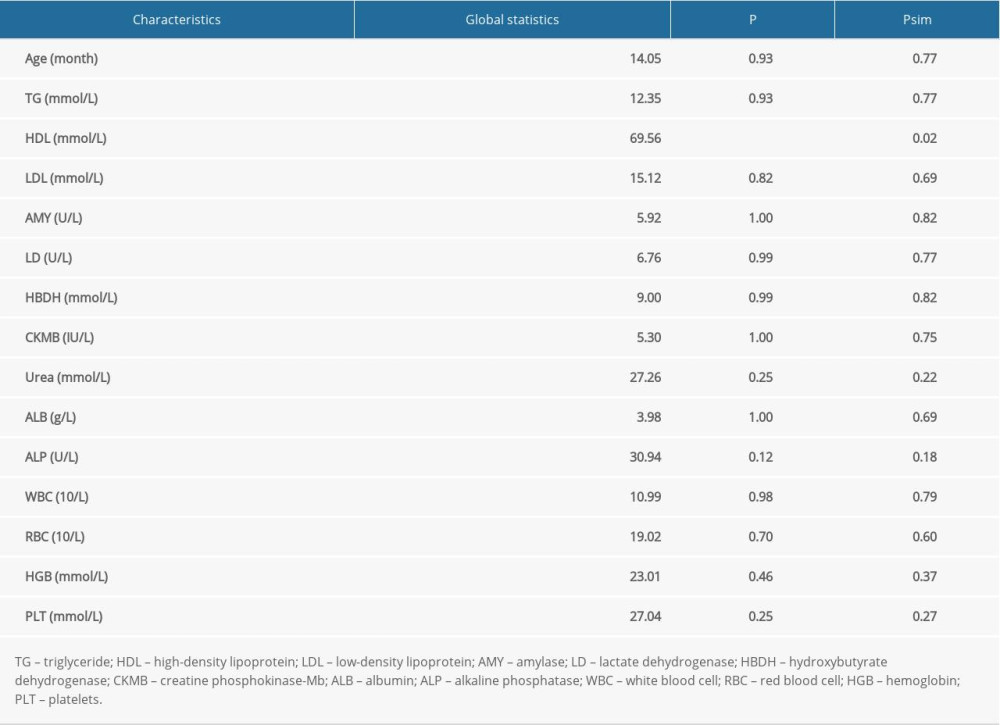 File 1. DNA extraction procedure.
File 1. DNA extraction procedure. File 2. Primers involved in the experiment.
File 2. Primers involved in the experiment.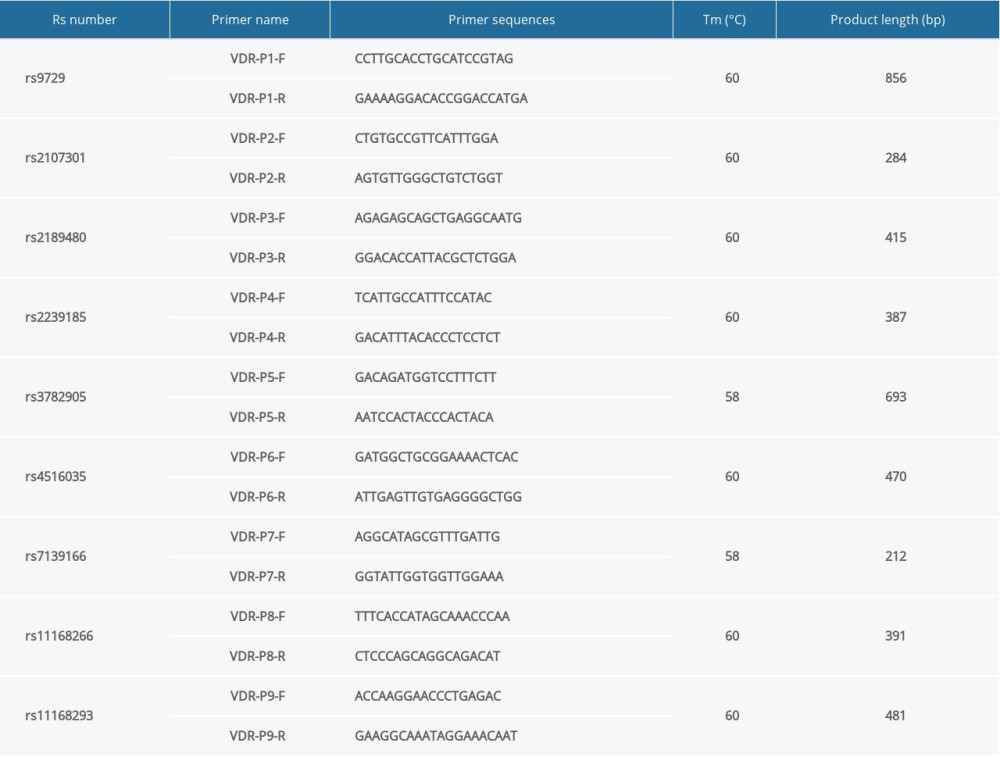
References
1. Weiss SL, Fitzgerald JC, Pappachan J, Global epidemiology of pediatric severe sepsis: The sepsis prevalence, outcomes, and therapies study: Am J Respir Crit Care Med, 2015; 191(10); 1147-57
2. Schlapbach LJ, Kissoon N, Defining pediatric sepsis: JAMA Pediatr, 2018; 172(4); 312-14
3. Chen F, Wang Y, Zhang W, A functional polymorphism-mediated disruption of EGR1/ADAM10 pathway confers the risk of sepsis progression: mBio, 2019; 10(4); e01663-19
4. Dahmer MK, Randolph A, Vitali S, Genetic polymorphisms in sepsis: Pediatr Crit Care Med, 2005; 6(3 Suppl); S61-73
5. Bizzarro MJ, Jiang Y, Hussain N, The impact of environmental and genetic factors on neonatal late-onset sepsis: J Pediatr, 2011; 158(2); 234-8e1
6. Zhu M, Zhao S, Candidate gene identification approach: progress and challenges: Int J Biol Sci, 2007; 3(7); 420-27
7. Yue LL, Wang FC, Zhang ML, Association of ATM and BMI-1 genetic variation with breast cancer risk in Han Chinese: J Cell Mol Med, 2018; 22(7); 3671-78
8. Tayel SI, Soliman SE, Elsayed HM, Vitamin D deficiency and vitamin D receptor variants in mothers and their neonates are risk factors for neonatal sepsis: Steroids, 2018; 134; 37-42
9. Bakke D, Sun J, Ancient nuclear receptor VDR with new functions: Microbiome and inflammation: Inflamm Bowel Dis, 2018; 24(6); 1149-54
10. Wang Y, Zhu J, DeLuca HF, Where is the vitamin D receptor?: Arch Biochem Biophys, 2012; 523(1); 123-33
11. Du J, Jiang S, Hu Z, Vitamin D receptor activation protects against lipopolysaccharide-induced acute kidney injury through suppression of tubular cell apoptosis: Am J Physiol Renal Physiol, 2019; 316(5); F1068-77
12. Kong J, Zhu X, Shi Y, VDR attenuates acute lung injury by blocking Ang-2-Tie-2 pathway and renin-angiotensin system: Mol Endocrinol, 2013; 27(12); 2116-25
13. Shang S, Wu J, Li X, Artesunate interacts with the vitamin D receptor to reverse sepsis-induced immunosuppression in a mouse model via enhancing autophagy: Br J Pharmacol, 2020; 177(18); 4147-65
14. Goldstein B, Giroir B, Randolph A, International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics: Pediatr Crit Care Med, 2005; 6(1); 2-8
15. Ban Y, Taniyama M, Ban Y, Vitamin D receptor gene polymorphism is associated with Graves’ disease in the Japanese population: J Clin Endocrinol Metab, 2000; 85(12); 4639-43
16. Kaya-Akyüzlü D, Kayaaltı Z, Söylemez E, Does maternal VDR FokI single nucleotide polymorphism have an effect on lead levels of placenta, maternal and cord bloods?: Placenta, 2015; 36(8); 870-75
17. Meyer V, Bornman L, Cdx-2 polymorphism in the vitamin D receptor gene (VDR) marks VDR expression in monocyte/macrophages through VDR promoter methylation: Immunogenetics, 2018; 70(8); 523-32
18. Yang Z, Wang Q, Zhong JF, Polymorphisms of the VDR gene in patients with nephrolithiasis in a Han Chinese population: Urolithiasis, 2019; 47(2); 149-54
19. Zhang L, Zhang S, He C, VDR gene polymorphisms and allergic diseases: Evidence from a meta-analysis: Immunol Invest, 2020; 49(1–2); 166-77
20. Zeljic K, Elkilany A, Supic G, Vitamin D receptor gene polymorphisms association with the risk of sepsis and mortality: Int J Immunogenet, 2017; 44(3); 129-34
21. Baralle D, Baralle M, Splicing in action: Assessing disease causing sequence changes: J Med Genet, 2005; 42(10); 737-48
22. Bouillon R, Carmeliet G, Verlinden L, Vitamin D and human health: Lessons from vitamin D receptor null mice: Endocr Rev, 2008; 29(6); 726-76
23. Zhao B, Xu N, Li R, Vitamin D/VDR signaling suppresses microRNA-802-induced apoptosis of keratinocytes in oral lichen planus: FASEB J, 2019; 33(1); 1042-50
24. Zgaga L, O;Sullivan F, Cantwell MM, Markers of vitamin D exposure and esophageal cancer risk: A systematic review and meta-analysis: Cancer Epidemiol Biomarkers Prev, 2016; 25(6); 877-86
25. Gu H, Wang X, Zheng L, Vitamin D receptor gene polymorphisms and esophageal cancer risk in a Chinese population: A negative study: Med Oncol, 2014; 31(2); 827
26. Shen F, Guo C, Wang Y, Low serum 25-hydroxyvitamin D levels may increase the detrimental effect of VDR variants on the risk of essential hypertension: Eur J Clin Nutr, 2020; 74(7); 1091-99
27. El Khoudary SR, HDL and the menopause: Curr Opin Lipidol, 2017; 28(4); 328-36
28. Gordon DJ, Rifkind BM, High-density lipoprotein – the clinical implications of recent studies: N Engl J Med, 1989; 321(19); 1311-16
29. Kajani S, Curley S, McGillicuddy FC, Unravelling HDL-looking beyond the cholesterol surface to the quality within: Int J Mol Sci, 2018; 19(7); 1971
30. Al-Daghri NM, Al-Attas OS, Alkharfy KM, Association of VDR-gene variants with factors related to the metabolic syndrome, type 2 diabetes and vitamin D deficiency: Gene, 2014; 542(2); 129-33
31. Gussago C, Arosio B, Guerini FR, Impact of vitamin D receptor polymorphisms in centenarians: Endocrine, 2016; 53(2); 558-64
32. Sun H, Long SR, Li X, Serum vitamin D deficiency and vitamin D receptor gene polymorphism are associated with increased risk of cardiovascular disease in a Chinese rural population: Nutr Res, 2019; 61; 13-21
33. Shen F, Wang Y, Sun H, Vitamin D receptor gene polymorphisms are associated with triceps skin fold thickness and body fat percentage but not with body mass index or waist circumference in Han Chinese: Lipids Health Dis, 2019; 18(1); 97
34. Xu Y, Lou Y, Kong J, VDR regulates energy metabolism by modulating remodeling in adipose tissue: Eur J Pharmacol, 2019; 865; 172761
35. Matthews DG, D’Angelo J, Drelich J, Adipose-specific VDR deletion alters body fat and enhances mammary epithelial density: J Steroid Biochem Mol Biol, 2016; 164; 299-308
36. Juárez-Rojas JG, Torre-Villalvazo I, Medina-Urrutia AX, Participation of white adipose tissue dysfunction on circulating HDL cholesterol and HDL particle size in apparently healthy humans: Int J Obes (Lond), 2020; 44(4); 920-28
Tables
 Table 1. The baseline characteristics of study children.
Table 1. The baseline characteristics of study children. Table 2. Genotype and allele distributions of VDR gene 9 studied variants between cases and controls, and genotype-based risk prediction for sepsis mortality risk.
Table 2. Genotype and allele distributions of VDR gene 9 studied variants between cases and controls, and genotype-based risk prediction for sepsis mortality risk. Table 3. The unadjusted and adjusted risk prediction of 9 studied variants for sepsis mortality risk under additive and dominant models, respectively.
Table 3. The unadjusted and adjusted risk prediction of 9 studied variants for sepsis mortality risk under additive and dominant models, respectively. Table 4. Haplotype frequencies (>1% in all cases and controls) of variants in VDR genes between cases and controls, and haplotype-based risk prediction for sepsis mortality risk.
Table 4. Haplotype frequencies (>1% in all cases and controls) of variants in VDR genes between cases and controls, and haplotype-based risk prediction for sepsis mortality risk. Table 5. Global testing of all haplotypes with anthropometric index and clinical biomarkers.
Table 5. Global testing of all haplotypes with anthropometric index and clinical biomarkers. Table 1. The baseline characteristics of study children.
Table 1. The baseline characteristics of study children. Table 2. Genotype and allele distributions of VDR gene 9 studied variants between cases and controls, and genotype-based risk prediction for sepsis mortality risk.
Table 2. Genotype and allele distributions of VDR gene 9 studied variants between cases and controls, and genotype-based risk prediction for sepsis mortality risk. Table 3. The unadjusted and adjusted risk prediction of 9 studied variants for sepsis mortality risk under additive and dominant models, respectively.
Table 3. The unadjusted and adjusted risk prediction of 9 studied variants for sepsis mortality risk under additive and dominant models, respectively. Table 4. Haplotype frequencies (>1% in all cases and controls) of variants in VDR genes between cases and controls, and haplotype-based risk prediction for sepsis mortality risk.
Table 4. Haplotype frequencies (>1% in all cases and controls) of variants in VDR genes between cases and controls, and haplotype-based risk prediction for sepsis mortality risk. Table 5. Global testing of all haplotypes with anthropometric index and clinical biomarkers.
Table 5. Global testing of all haplotypes with anthropometric index and clinical biomarkers. File 1. DNA extraction procedure.
File 1. DNA extraction procedure. File 2. Primers involved in the experiment.
File 2. Primers involved in the experiment. In Press
06 Mar 2024 : Clinical Research
Comparison of Outcomes between Single-Level and Double-Level Corpectomy in Thoracolumbar Reconstruction: A ...Med Sci Monit In Press; DOI: 10.12659/MSM.943797
21 Mar 2024 : Meta-Analysis
Economic Evaluation of COVID-19 Screening Tests and Surveillance Strategies in Low-Income, Middle-Income, a...Med Sci Monit In Press; DOI: 10.12659/MSM.943863
10 Apr 2024 : Clinical Research
Predicting Acute Cardiovascular Complications in COVID-19: Insights from a Specialized Cardiac Referral Dep...Med Sci Monit In Press; DOI: 10.12659/MSM.942612
06 Mar 2024 : Clinical Research
Enhanced Surgical Outcomes of Popliteal Cyst Excision: A Retrospective Study Comparing Arthroscopic Debride...Med Sci Monit In Press; DOI: 10.12659/MSM.941102
Most Viewed Current Articles
17 Jan 2024 : Review article
Vaccination Guidelines for Pregnant Women: Addressing COVID-19 and the Omicron VariantDOI :10.12659/MSM.942799
Med Sci Monit 2024; 30:e942799
14 Dec 2022 : Clinical Research
Prevalence and Variability of Allergen-Specific Immunoglobulin E in Patients with Elevated Tryptase LevelsDOI :10.12659/MSM.937990
Med Sci Monit 2022; 28:e937990
16 May 2023 : Clinical Research
Electrophysiological Testing for an Auditory Processing Disorder and Reading Performance in 54 School Stude...DOI :10.12659/MSM.940387
Med Sci Monit 2023; 29:e940387
01 Jan 2022 : Editorial
Editorial: Current Status of Oral Antiviral Drug Treatments for SARS-CoV-2 Infection in Non-Hospitalized Pa...DOI :10.12659/MSM.935952
Med Sci Monit 2022; 28:e935952