24 November 2022: Human Study
FLT3 Gene Mutations in Acute Myeloid Leukemia Patients in Northeast Thailand
Nang Ei Ei Aung1BCD, Supawadee Yamsri2ACDE*, Nattiya Teawtrakul3B, Piyawan Kamsaen4B, Supan FucharoenDOI: 10.12659/MSMBR.937446
Med Sci Monit Basic Res 2022; 28:e937446
Abstract
BACKGROUND: Mutations in the FLT3 gene are associated with acute myeloid leukemia (AML). FLT3 mutations have been identified in approximately 30% of de novo AML patients, particularly those with typical karyotype and inferior prognosis. Therefore, we examined the frequencies of an internal tandem duplication (ITD) and missense mutations of the FLT3 gene and their associated clinical characteristics in patients with AML in northeast Thailand.
MATERIAL AND METHODS: The leftover bone marrow and/or peripheral blood specimens of 65 newly diagnosed AML patients recruited from Srinagarind Hospital, Khon Kaen University, northeast Thailand, between January 2020 and May 2021 were included in this study. FLT3-ITD and FLT3- tyrosine kinase domain (TKD) were amplified using PCR-related techniques.
RESULTS: The frequencies of FLT3-ITD and TKD were found to be 16.9% (11/65) and 10.8% (7/65), respectively. One patient had ITD and TKD mutations. The white blood cell count and peripheral blast percentage of FLT3-ITD-positive patients were statistically significantly higher than those of the FLT3-wild type patients, while the peripheral blast percentage of FLT3-TKD-positive patients was significantly lower. No other clinical characteristics among FLT3-positive and FLT3-wild-type patients were observed. DNA sequencing identified 4 FLT3-TKD mutations. The c.2504A>T; Asp835Val and c.2503G>C; Asp835His mutations were predicted as pathogenic mutations while the 2 novel mutations, c.2508C>A; Ile836= and c.2508C>G; Ile836Met were predicted as neutral mutations.
CONCLUSIONS: This study showed for the first time that FLT3-TKD mutation is common among northeast Thai AML patients. The data should prove useful for selecting efficacious targeted treatment plans for the patients.
Keywords: flt3 Ligand Protein, Leukemia, Myeloid, Acute, Humans, Thailand, Mutation, Prognosis, Treatment Outcome, fms-Like Tyrosine Kinase 3
Background
Acute myeloid leukemia (AML) is caused by genetic and epigenetic alterations of hematopoietic stem cells or progenitor cells. The lesions revise normal hematopoietic growth and differentiation into the proliferation of immature, atypical myeloid progenitor cells in the bone marrow and peripheral blood [1]. Cytogenetics results are well-known prognostic markers that influence patient prognosis and therapy [2,3]. However, cytogenetics reveals a normal karyotype in about 50% of patients with AML [3,4]. A spectrum of molecular mutation plays a critical part in the disease’s pathogenesis and is important for determining the prognosis of patients with AML. For example, NPM1 and CEBPA are favorable prognostic markers [5–7], while FLT3-ITD and MLL-PTD are associated with poor prognosis [5,6].
One of the most frequent genetic abnormalities found in AML is FMS-like tyrosine kinase 3 (FLT3) gene mutation. The FLT3 genes belong to the tyrosine kinase class III receptors on chromosome 13 at band q12 and encompass 24 exons [7]. The FLT3 receptors consist of 5 immunoglobulin-like extracellular ligand-binding regions, a single transmembrane region, and an intra-cytoplasmic region that supports the 2 tyrosine kinase domains (TKDs, TKD1 and TKD2) and the juxtamembrane domain [7,8]. FLT3 mutations in AML have been identified in approximately one-third of newly diagnosed AML cases, especially those with a normal karyotype [8]. Two distinct types of activating mutations in the FLT3 gene have been reported: internal tandem duplication in the juxtamembrane domain (FLT3-ITD) and missense mutations in the TKD (FLT3-TKD) [9]. The FLT3-ITD mutation is the most common in patients with AML [8]. It is known as a poor prognostic factor and is associated with an increased risk of relapse, shortened disease-free survival, and inferior overall survival [10]. The FLT3-TKD mutation, despite its infrequency (approximately 7–10%) in AML as ITD, has a more complex prognosis, with reports of adverse effects, no effect, and favorable prognosis [9,11,12]. The emergence of FLT3-TKD mutations as an acquired resistance in FLT3-ITD-positive patients after treatment with FLT3 inhibitors is also a major concern in patients with AML [13,14]. Patients with AML who carry both ITD and TKD mutations have worse outcomes than patients with either TKD or ITD mutation alone. Therefore, the FLT3 receptor tyrosine kinase is a crucial biomarker of interest, and identifying both mutations is important as it can guide individualized treatment plans for patients with AML [15].
Studies of FLT3-ITD mutations in Thailand showed the frequency among adult AML patients ranging from 21% to 25% in central, northeast, and upper northern Thailand [16–18]. AML has a frequency of 6.3% to 14.3% in pediatric patients in northeast Thailand [16,19]. Variable frequencies of FLT3-ITD mutations have been reported in Malaysia (16.1%) [20], India (18.8%) [21], Taiwan (15.4%) [22], and China (25.9%) [23]. The frequency of FLT3-TKD was found to be 3.3% to 18.7% in Taiwan [22,24], 6.3% in China [23], 7% in Japan [25], and 4.8% to 11% in Europe [12,14,26,27]. In Thailand, a frequency of 3.1% of FLT3-TKD mutation has been reported among adult AML patients in central Thailand [17], whereas 4.8% were found in pediatric AML patients in northeast Thailand [19]. The 2 most common FLT3-TKD mutations, Asp835Tyr and Asp835His, were found in central Thailand. However, there is no information about the type of FLT3-TKD mutations found in northeast Thailand. Moreover, studies on FLT3-ITD and FLT3-TKD mutations in northeast Thailand are scarce. Therefore, in this study, we investigated the frequencies of FLT3-ITD and FLT3-TKD mutations and the patient characteristics among northeast Thai patients with AML.
Material and Methods
PATIENT SPECIMENS:
Leftover blood or bone marrow specimens were selectively studied prospectively from 65 newly diagnosed AML patients recruited from Srinagarind Hospital, Khon Kaen University, northeast Thailand. The study included all patients newly diagnosed with AML between January 2020 and May 2021 at Srinagarind Hospital. The diagnosis of AML was made according to the French-American-British classification. Complete blood counts and morphology were performed in all patients. To confirm the AML, a bone marrow examination was done. Immunophenotyping was performed by flow cytometry using bone marrow specimens or peripheral blood specimens. The diagnosis of AML was classified into 2 groups, AML-M3 and AML-nonM3, according to the national guideline for treating AML. The inclusion criteria were patients with a diagnosis of AML by flow cytometry. DNA was extracted from all samples using the GF-1 Blood DNA Extraction kit (Vivantis Technologies, Malaysia), as described in the manufacturer’s protocol. This study was approved by the Institutional Review Board of Khon Kaen University (HE631400), Khon Kaen, Thailand. The study was registered in the Thai Clinical Trails Registry (registration no. TCTR20220928006).
ANALYSIS OF FLT3-ITD MUTATION:
FLT3-ITD mutation was identified using the conventional PCR technique from Kiyoi et al in 1997 [28]. The primers were designed to specifically amplify both the FLT3-ITD mutant and wild-type FLT3 at exons 14 and 15. The forward primer, 11F, (5′-GCAATTTAGGTATGAAAGCCAGC-3′) was used with the reverse primer, 12R, (5′-CTTTCAGCATTTTGACGGCAACC-3′) to produce a 329 bp wild-type product. Any patient with an additional PCR product longer than 329 bp was considered to have FLT3-ITD mutant type. The PCR reaction was composed of 50 ng of genomic DNA, 200 μM dNTPs, 10 pmol of each primer, and 1 unit of Taq DNA polymerase (Biotools, B&M Labs, Madrid, Spain). Denaturing, annealing, and extension steps were conducted at 94°C for 3 min, followed by 30 cycles of 93°C for 30 s, 60°C for 30 s, and 72°C for 1 min in a SimpliAmp Thermal Cycler (Applied Biosystems, Waltham, MA, USA). The PCR amplicons were then analyzed on 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized under ultraviolet illumination. The presence of FLT3-ITD mutation was visualized as a DNA fragment larger than a wild-type gene fragment of 329 bp. The locations and orientations of primers are shown in Figure 1A.
IDENTIFICATION OF FLT3-TKD MUTATIONS:
Identification of FLT3-TKD mutations was done by using the PCR-restriction fragment length polymorphism (RFLP) assay. The newly designed primers corresponding to the DNA sequence in exon 20 were made, including the forward primer, F1, (5′-AGAACTGCAGCCACCATAGC-3′) and the reverse primer, R1, (5′-AGCCCAAGGACAGATGTGA-3′). The PCR reaction was composed of 50 ng of DNA, 200 μM dNTPs, 30 pmol of each primer, and 1 unit of Taq DNA polymerase (New England Biolab, Inc., USA). After initial heating at 94°C for 3 min, 30 cycles of 93°C for 30 s, 60°C for 30 s, and 72°C for 1 min was done in a SimpliAmp Thermal Cycler (Applied Biosystems). The amplified products were digested with EcoRV restriction endonuclease (New England Biolab, Inc., USA). The digestion products were separated on 2% agarose gel. In the presence of wild-type FLT3-TKD, the amplified products were digested into 2 fragments (295 bp and 260 bp), while undigested products having 555 bp indicated the presence of a FLT3-TKD mutation, as shown in Figure 2A. All FLT3-TKD mutations detected were verified by DNA sequencing.
DNA SEQUENCING:
All samples positive for FLT3-TKD mutations and 5 additional wild-type samples were confirmed by DNA sequencing. PCR products were purified using a GenepHlow gel/PCR kit (Geneaid Biotech Ltd., Taipei, Taiwan). Approximately 100 ng purified PCR products were directly sequenced using Sanger sequencing techniques (First BASE Lab, Selangor, Malaysia). Sequence analysis was done using a Seq-scanner program (Thermo Fisher Scientific, Waltham, MA, USA).
FUNCTIONAL PREDICTION OF THE FLT3-TKD MUTATIONS:
The functions of FLT3-TKD mutations were predicted using the Catalogue of Somatic Mutations in Cancer (COSMIC) database (https://cancer.sanger.ac.uk) [29]. The functional scores for individual mutations were reported using the Functional Analysis through Hidden Markov Models (FATHMM) prediction. FATHMM was done using a high-throughput web server to predict the functional consequences of coding and non-coding variants in the human genome. The functional scores range from 0 to 1. To highlight the most significant data in COSMIC, only scores ≥0.7 are classified as “pathogenic”. In contrast, mutations are classified as “neutral” if the score is ≤0.5.
STATISTICAL ANALYSIS:
The prevalence of FLT3-ITD and TKD mutations were summarized as a percentage with a 95% confidence interval (CI). The data were presented as medians and interquartile ranges (IQRs) for continuous variables. For comparison of continuous variables, such as age, white blood cell (WBC) count, blast count, hemoglobin level, and platelet count, the Mann-Whitney U test was used, using SPSS (version 25; IBM Corp, Armonk, NY, USA). Fisher’s exact and chi-square tests were used to analyze the differences in categorical variables. A P value <0.05 was considered statistically significant.
Results
The 2 PCR assays were successfully used to identify FLT3-ITD and FLT3-TKD mutations. The representative agarose gel electrophoresis of the PCR assay to detect FLT3-ITD is shown in Figure 1B. The wild-type FLT3-ITD showed 329 bp fragments. A fragment larger than 329 bp was observed in FLT3-ITD-positive patients. The molecular size of FLT3-ITD mutation was not examined in this study. Among 65 patients with AML, FLT3-ITD mutation was observed in 11 patients. Figure 2B shows a representative agarose gel electrophoresis of the PCR-RFLP assay for the detection of FLT3-TKD mutations. The 555-bp fragment of patients with wild-type FLT3-TKD was digested with EcoRV to produce the 260-bp and 295-bp fragments. In the FLT3-TKD-positive AML patients, the restriction site was modified by the mutation; therefore, only the 555-bp fragment was observed due to there being no digestion by EcoRV. A total of 7 patients with AML were positive for the FLT3-TKD mutation.
Among the 65 patients, the overall frequency of FLT3 mutation was 27.8% (17/65). The frequencies of FLT3-ITD and FLT3-TKD mutations were 16.9% (11/65) and 10.8% (7/65), respectively. One patient was a compound heterozygote for FLT3-ITD and TKD mutations (1.54%). When we divided the AML patients into adults and children, the frequencies of FLT3 mutations were 28.6% (14/49) and 25.0% (4/16), respectively. In adult AML patients, the frequencies of FLT3-ITD, FLT3-TKD, and compound FLT3 mutations were 18.4%, 10.2%, and 2.0%, respectively. In children, the frequencies of FLT3-ITD and FLT3-TKD mutations were 12.5% and 12.5%, respectively.
The characteristics and laboratory data of patients with newly diagnosed AML are summarized in Table 1. Eighteen male and 47 female patients were recruited (median age, 43 years). Among these 65 patients, 49 were adults (>18 years) and 16 were children (≤18 years). FLT3mutations were identified in 17 patients, including 10 patients with FLT3-ITD, 6 patients with FLT3-TKD, and 1 patient with both FLT3-ITD and FLT3-TKD mutations. The presence of FLT3-TKD mutations was related to neither sex nor age. The median WBC count in patients with wild-type FLT3 was 14.7×103/μL (IQR, 4.9–24.3). The median WBC count in all FLT3-positive patients, FLT3-ITD-positive patients, and FLT3-TKD-positive patients were 34.0 (IQR, 13.3–193.3), 90.3 (IQR, 26.6–535.0), and 17.3 (IQR, 8.3–75.4)×103/μL, respectively. A statistically significant difference was observed in all FLT3-positive patients and FLT3-ITD-positive patients compared with wild-type-FLT3 patients (
In this study, we found 7 patients with FLT3-TKD mutations. Further characterization was done by DNA sequencing. This DNA sequencing identified 4 different point mutations in codons 835 and 836 of the FLT3-TKD gene, as shown in Table 2. Four patients showed A to T nucleotide substitution at codon 835, which resulted in an amino acid substitution of aspartic acid to valine (c.2504A>T; Asp835Val). The 3 remaining patients had, respectively, Asp835His (c.2503G>C) and 2 novel mutations in codon 836, Ile836=(c.2508C>A) and Ile836Met (c.2508C>G) mutations. The DNA sequencing of these novel mutations is shown in Figure 3. The COSMIC database [29] was used to explore the effect of these mutations in patients with AML. FATHMM prediction showed that c.2504A>T and c.2503G>C were pathogenic mutations, with a functional score of 0.99. While, the novel mutations, c.2508C>A and c.2508C>G, were classified as neutral, with functional scores of 0.11 and 0.20, respectively.
Discussion
Activating mutations in the FLT3 gene are the most widely recognized genetic changes in patients with AML, occurring in approximately one-third of patients. The mutations in FLT3 generate structural changes in receptors and cause permanent tyrosine kinase activation in the absence of ligand, leading to leukemogenesis. It is known that FLT3 gene mutations are related to inferior prognosis. The FLT3 mutation is an important molecular target both at de novo and relapse for clinical prognosis and in selecting specific therapies against FLT3 receptor activation in patients with AML [14,16]. FLT3-ITD and FLT3-TKD mutations are commonly found in patients with AML. The most common type is the FLT3-ITD mutation. Identifying FLT3-ITD and FLT3-TKD mutations might provide useful information for selecting the proper treatment of AML. Therefore, we focused on the frequency of both mutations in our population and the association with patients’ characteristics. In our study, the FLT3-ITD mutation was found in 18.4% of adult AML patients, which is comparable to the result of the previous study in northeast Thailand by Kumsaen P et al (21.1%) [16]. Table 3 shows that FLT3-ITD was found to be common in adult AML patients in many studies [30–32]. The frequency of FLT3-ITD has been reported in central and northern Thailand, with a frequency 24.6% and 25.0%, respectively [17,18]. In Asian countries, frequencies of 19.0% to 25.9% were reported in Malaysia, India, China, Taiwan, and Japan [20–23,25]. This indicates that the FLT3-ITD mutation is common in adult AML patients. The frequency of FLT3-ITD among pediatric AML patients was found to be 12.5% in the present study, which is different from the previous study in northeast Thailand (4.8%) [19]. This might be due to the difference in age of the studied samples. The median (range) age in this study was 15 (2–17) years old, while the previous study was 10 (6–12) years old. According to the patients’ characteristics, we found that FLT3-ITD-positive patients had a higher WBC count, percentage of peripheral blood blasts, and percentage of bone marrow blasts as compared with those with wild-type FLT3. These findings are consistent with other studies in Thailand [16,17,19]. In the presence of ITD, the juxtamembrane domain undergoes a conformational change, losing its autoinhibitory function, which causes the subsequent activation of the receptor and downstream signaling pathways, including the Ras/MAPK pathway and PI3K/Akt pathway. These result in a high proliferation of WBC and blast cells [33,34]. Most patients with AML in the present study received standard treatment, the 7+3 regimen. However, the rates of complete remission varied. Unfortunately, the treatment patterns and outcomes were unobtainable in this study.
FLT3-TKD mutation has been found in adult and pediatric AML patients with variable frequency in different populations [20–23,25]. Our study showed the frequency of FLT3-TKD as 10.2% and 12.5% among adult and pediatric patients with AML, respectively, which differs from that previously reported in central Thailand (3.1%) [17]. This data might suggest that the FLT3-TKD mutation is more common in northeast Thailand. This discrepancy might be due to differences in ethnic background, population genetics, and environmental factors. Another report among pediatric AML patients in northeast Thailand showed a lower frequency of FLT3-TKD mutation, as compared with that of our study, which could be due to the difference of patient age, as described previously. A previous study from Taiwan showed that the FLT3-TKD mutation is common in AML-M3 [22]. In our study, among 7 patients with AML-M3, 2 patients (40%) were positive for FLT3-TKD. While among 41 patients with AML-nonM3, only 5 patients (9.8%) were positive for FLT3-TKD. In other studies, the frequency of FLT3-TKD mutations were found to be 4.8% to 7.7% in Germany [14,27], 11% in the United Kingdom [12], and 10.1% in Sweden [26]. Furthermore, in the present study, healthy individuals were also analyzed to exclude the possibility of polymorphisms. FLT3-TKD mutations were not found in all healthy individuals (data not shown). These results confirmed that FLT3-TKD mutations were associated with the molecular pathogenesis of AML. No association was seen between age or sex and the presence of FLT3-TKD mutations. FLT3-TKD-positive AML patients in this study were associated with lower peripheral blood and bone marrow blast percentages when compared with FLT3-ITD-positive patients, which was consistent with the previous report [35]. However, another study did not find this result [20,22]. Functionally, both FLT3 mutations induce activation of the signaling pathways without ligand binding to the receptor. This unregulated activation impairs normal hematopoietic growth and differentiation into the accumulation of leukemic blast and immature myeloid cells in the peripheral blood and bone marrow [14]. Variations in the biological characteristics of the patients, such as cytogenetics (whole AML or AML with normal karyotype), might contribute to the heterogeneity of the results. However, in the present study, the cytogenetics was not available in all patients. Therefore, the association of clinicohematological parameters according to cytogenetics could not be determined.
Types of FLT3-TKD mutations among patients with AML in northeast Thailand were reported here for the first time. DNA sequencing of 7 FLT3-TKD-positive AML patients showed 4 different mutations. There are 2 mutations in codon 835, including 835 GAT – GTT (c.2504A>T; Asp835Val) and 835 GAT – CAT (c.2503G>C; p.Asp835His) that were previously found in central Thai populations. The 2 mutations in codon 836 included 836 ATC – ATA (c.2508C>A; p.Ile836=) and 836 ATC – ATG (c.2508 C>G; p.Ile836Met) and were found for the first time in a Thai population. In the present study, the most frequent mutation was Asp835Val, while in the central Thai population, the most common mutation was found to be Asp835Tyr [17]. This indicates that the type of FLT3-TKD mutation is varied among populations.
In this study, we found that 2 mutations in codon 835 (c.2504A>T and c.2503G>C) were classified as pathogenic, with high functional scores (0.99). The COSMIC database [29] also showed that c.2504A>T and c.2503G>C are resisted to the drugs quizartinib and sorafenib, respectively. Quizartinib and sorafenib are second-generation tyrosine kinase inhibitors (TKIs). Recently, many TKIs have been used with standard protocols in adult AML patients. However, it was demonstrated that these 2 mutations in codon 835 of the FLT3 gene are important and have a pathological effect on AML. In the present study, 5 of 7 patients (71.4%) carried these mutations. Therefore, identification of these mutations among patients with AML is essential. The other 2 novel mutations in codon 836 (c.2508C>A and c.2508 C>G) are classified as neutral and have no effect on AML patients. Likely, the pathogenic effects of c.2504A>T and c.2503G>C mutations involve the activation loop of the TK2 domain, primarily restricted to the carboxy-terminal portion of the catalytic domain, resulting to autophosphorylated and activate known downstream targets of FLT3 [32,36]. Therefore, it has been demonstrated that younger adults with FLT3-ITD-positive de novo AML who have FLT3-TKD mutations in D835/I836 are associated with worse disease-free survival than that of similar patients without FLT3
In this study, 1 patient had compound heterozygous FLT3-ITD and FLT3-TKD mutations. The patient was a 59-year-old woman. She had a WBC count of 89.4×103/μL, Hb level of 7.8 g/dL, and platelet count of 38×109/μL, and the number of peripheral blood blasts was 93%. Her overall survival was 11 days after diagnosis. The elevated WBC count and high number of peripheral blood blasts showed the typical characteristics of FLT3-ITD-positive AML patients. However, the prognostic significance of FLT3 mutations in this patient is unclear. Further study should be done. However, this data support other studies that showed FLT3-ITD and FLT3-TKD had a low coexistence [17,25,27]. In addition to FLT3-ITD and TKD mutations, other mutations, such as NPM1, MLL-PTD, and CEBPA, were associated with AML patient prognosis [14,17,25,27]. However, data on these mutations among patients with AML in northeast Thailand is still limited. Although the French-American-British classification system is useful for classifying AML patients, many factors affecting the prognosis are not taken into consideration. Therefore, the WHO classification system is alternatively recommended. Subsequently, further study of other molecular factors is essential for better understanding of the molecular pathogenesis of AML patients in the region.
Conclusions
The frequency of FLT3-ITD mutation is relatively high in patients in northeast Thailand, while the prevalence of FLT3-TKD mutation in northeast Thailand was as high as in other studies. Therefore, laboratory investigations of these 2 mutations should be performed routinely in all patients at the diagnosis of AML to provide targeted therapies. However, further study on the impact of the type of FLT3-TKD mutation with the proper FLT3 TKI in patients with AML is required.
In addition, the characterization of FLT3-TKD mutations is suggested for selecting the proper FLT3 TKI. However, further study on the prognostic impact of FLT3-TKD mutations among AML patients is required.
Figures
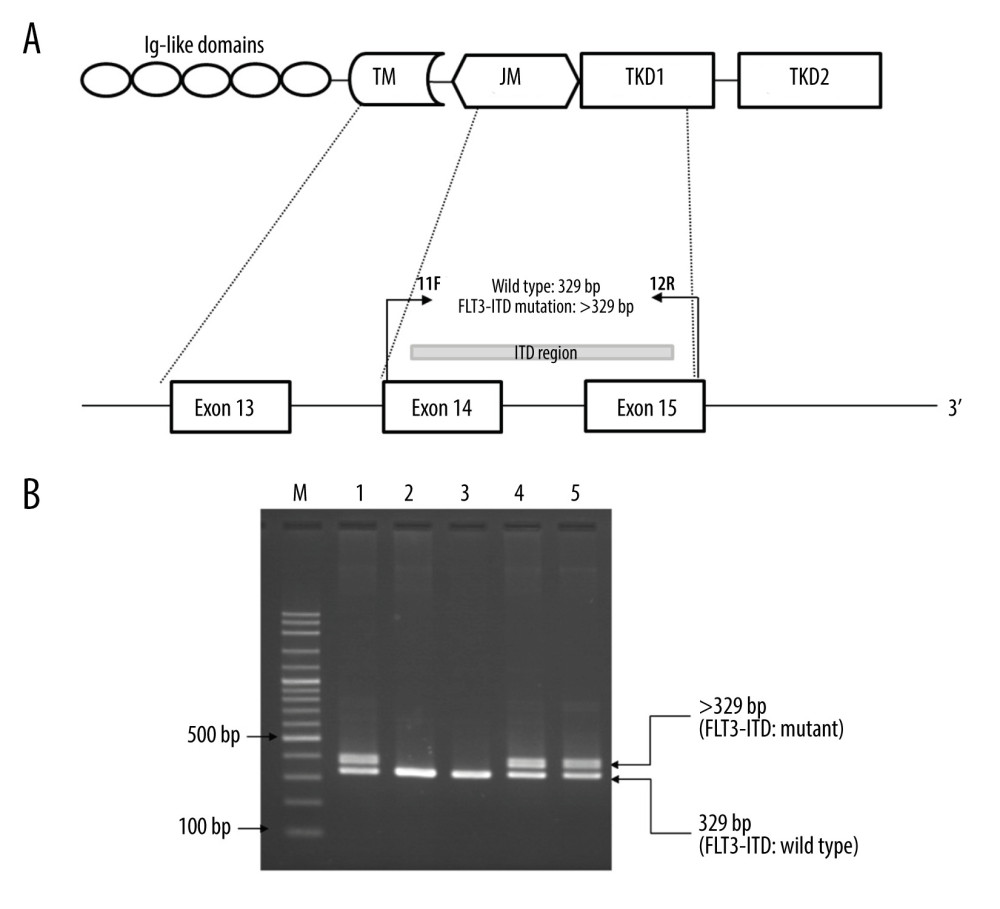 Figure 1. Identification of FMS-like tyrosine kinase 3 (FLT3)-internal tandem duplication (ITD) mutation by PCR. (A) Locations and orientations of primers used for identification of FLT3-ITD mutation. Primers 11F and 12R were used to amplify FLT3-ITD mutant and FLT3-wild type at exons 14–15. The 329 bp fragment indicates the wild type while the fragment larger than 329 bp indicates the FLT3-ITD mutation. TM – transmembrane domain; JM – juxtamembrane domain; TKD – tyrosine kinase domain. (B) A representative 2% agarose gel electrophoresis of PCR products. Lane 1 and lane 2 are positive and negative control for the FLT3-ITD mutation, respectively. Lanes 3–5 are PCR products of patients with acute myeloid leukemia. Lane 3 shows the 329 bp fragment indicating the wild type. Lane 4 & 5 show the 329 bp and the fragment larger than 329 bp (>329) indicating FLT3-ITD mutation. M indicates 100 bp DNA ladder.
Figure 1. Identification of FMS-like tyrosine kinase 3 (FLT3)-internal tandem duplication (ITD) mutation by PCR. (A) Locations and orientations of primers used for identification of FLT3-ITD mutation. Primers 11F and 12R were used to amplify FLT3-ITD mutant and FLT3-wild type at exons 14–15. The 329 bp fragment indicates the wild type while the fragment larger than 329 bp indicates the FLT3-ITD mutation. TM – transmembrane domain; JM – juxtamembrane domain; TKD – tyrosine kinase domain. (B) A representative 2% agarose gel electrophoresis of PCR products. Lane 1 and lane 2 are positive and negative control for the FLT3-ITD mutation, respectively. Lanes 3–5 are PCR products of patients with acute myeloid leukemia. Lane 3 shows the 329 bp fragment indicating the wild type. Lane 4 & 5 show the 329 bp and the fragment larger than 329 bp (>329) indicating FLT3-ITD mutation. M indicates 100 bp DNA ladder. 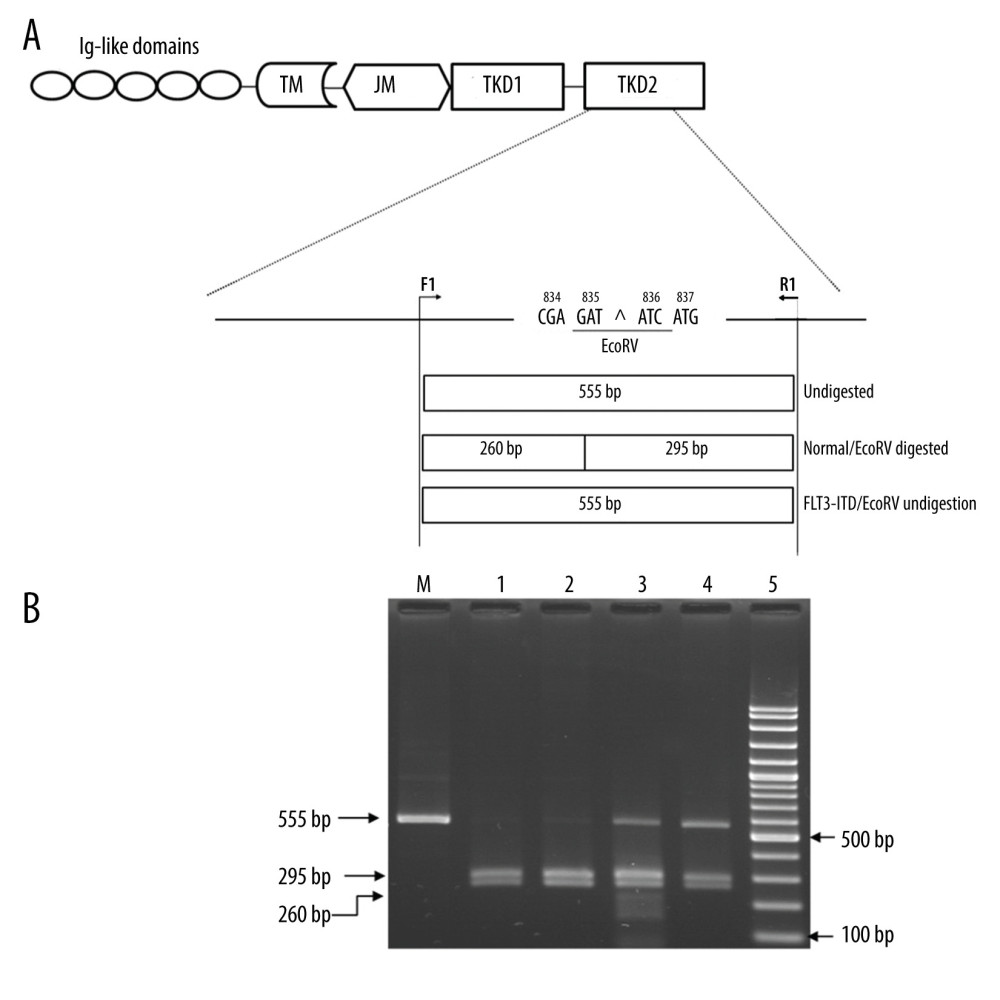 Figure 2. Identification of FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutation by PCR technique followed by EcoRV digestion. (A) Locations and orientations of primers. Forward primer, F1, and reverse primer, R1, specifically to codon 20 of TKD2 domain were used to produce a 555 bp fragment. Codons 835 and 836 are encoded by nucleotides GATATC which form the EcoRV restriction site. The PCR product of wild type patients were digested by EcoRV into 2 fragments, 260 and 295 bp fragments. The acute myeloid leukemia patients with FLT3-TKD mutations, the PCR product shows 555 bp undigested fragment. TM – transmembrane domain; JM – juxtamembrane domain; TKD – tyrosine kinase domain. (B) A representative 2% agarose gel electrophoresis of PCR products among AML patients after EcoRV digestion. Lane 1 shows uncut PCR Product. Lane 2 & 3, wild type patients show PCR-RFLP fragments which the EcoRV digested produced 260 and 295 bp fragments. Lanes 4 and 5 are positive for FLT3-internal tandem duplication (ITD) mutation which show 555 bp undigested fragments. M indicates the 100 bp DNA ladder.
Figure 2. Identification of FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutation by PCR technique followed by EcoRV digestion. (A) Locations and orientations of primers. Forward primer, F1, and reverse primer, R1, specifically to codon 20 of TKD2 domain were used to produce a 555 bp fragment. Codons 835 and 836 are encoded by nucleotides GATATC which form the EcoRV restriction site. The PCR product of wild type patients were digested by EcoRV into 2 fragments, 260 and 295 bp fragments. The acute myeloid leukemia patients with FLT3-TKD mutations, the PCR product shows 555 bp undigested fragment. TM – transmembrane domain; JM – juxtamembrane domain; TKD – tyrosine kinase domain. (B) A representative 2% agarose gel electrophoresis of PCR products among AML patients after EcoRV digestion. Lane 1 shows uncut PCR Product. Lane 2 & 3, wild type patients show PCR-RFLP fragments which the EcoRV digested produced 260 and 295 bp fragments. Lanes 4 and 5 are positive for FLT3-internal tandem duplication (ITD) mutation which show 555 bp undigested fragments. M indicates the 100 bp DNA ladder. 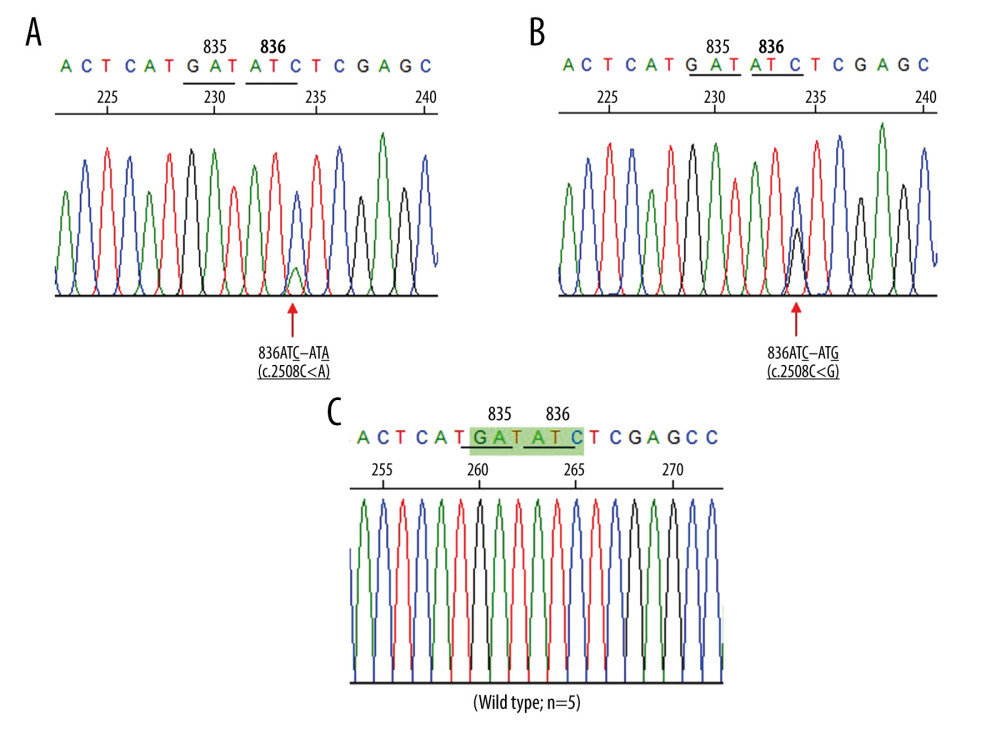 Figure 3. (A, B) Direct sequencing in the forward direction demonstrating 2 novel FMS-like tyrosine kinase 3 (FLT3)-TKD mutations among AML patients. Red arrow indicates the mutation site. The mutation and 836ATC – ATG mutation, respectively. (C) The corresponding DNA sequence of AML patients with wild type FLT3-TKD at the same region.
Figure 3. (A, B) Direct sequencing in the forward direction demonstrating 2 novel FMS-like tyrosine kinase 3 (FLT3)-TKD mutations among AML patients. Red arrow indicates the mutation site. The mutation and 836ATC – ATG mutation, respectively. (C) The corresponding DNA sequence of AML patients with wild type FLT3-TKD at the same region. Tables
Table 1. Characteristics and laboratory data of 65 newly diagnosed acute myeloid leukemia (AML) patients according to type of FLt3 mutation.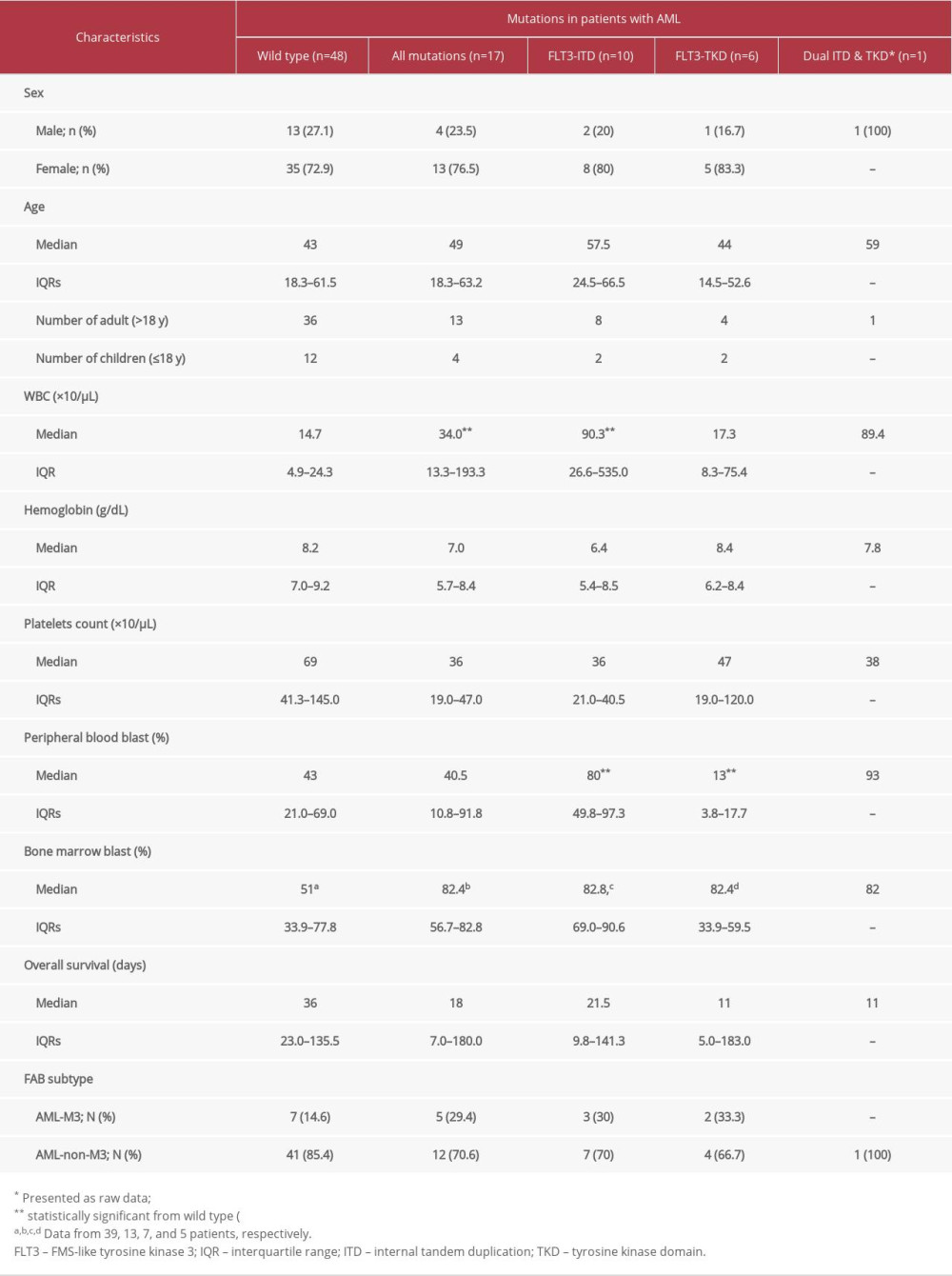 Table 2. Characteristics of patients with FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutations.
Table 2. Characteristics of patients with FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutations.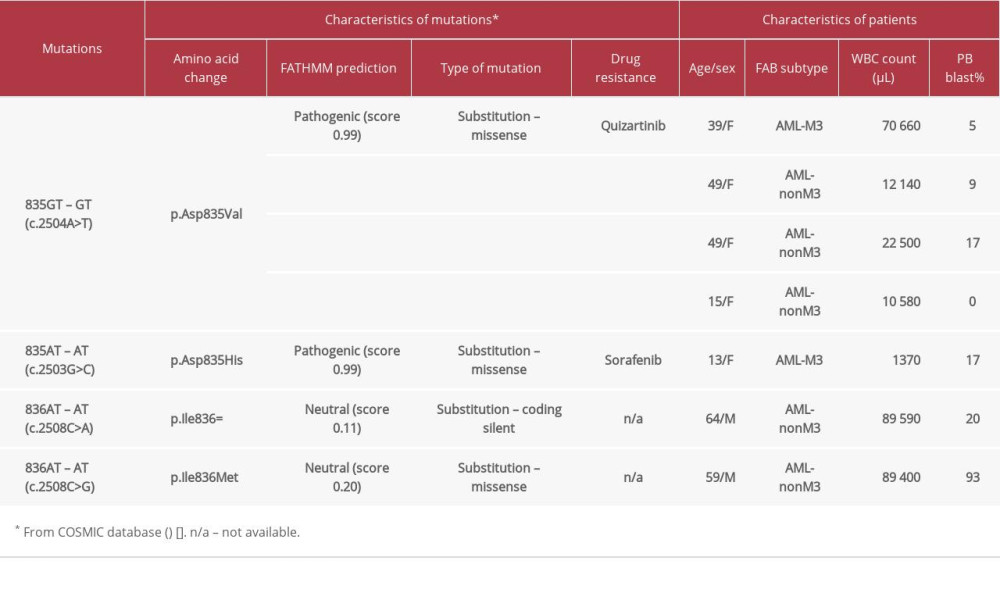 Table 3. Frequency of FMS-like tyrosine kinase 3 (FLT3)-ITD and tyrosine kinase domain (TKD) mutations in patients with acute myeloid leukemia (AML) from various studies in Asia.
Table 3. Frequency of FMS-like tyrosine kinase 3 (FLT3)-ITD and tyrosine kinase domain (TKD) mutations in patients with acute myeloid leukemia (AML) from various studies in Asia.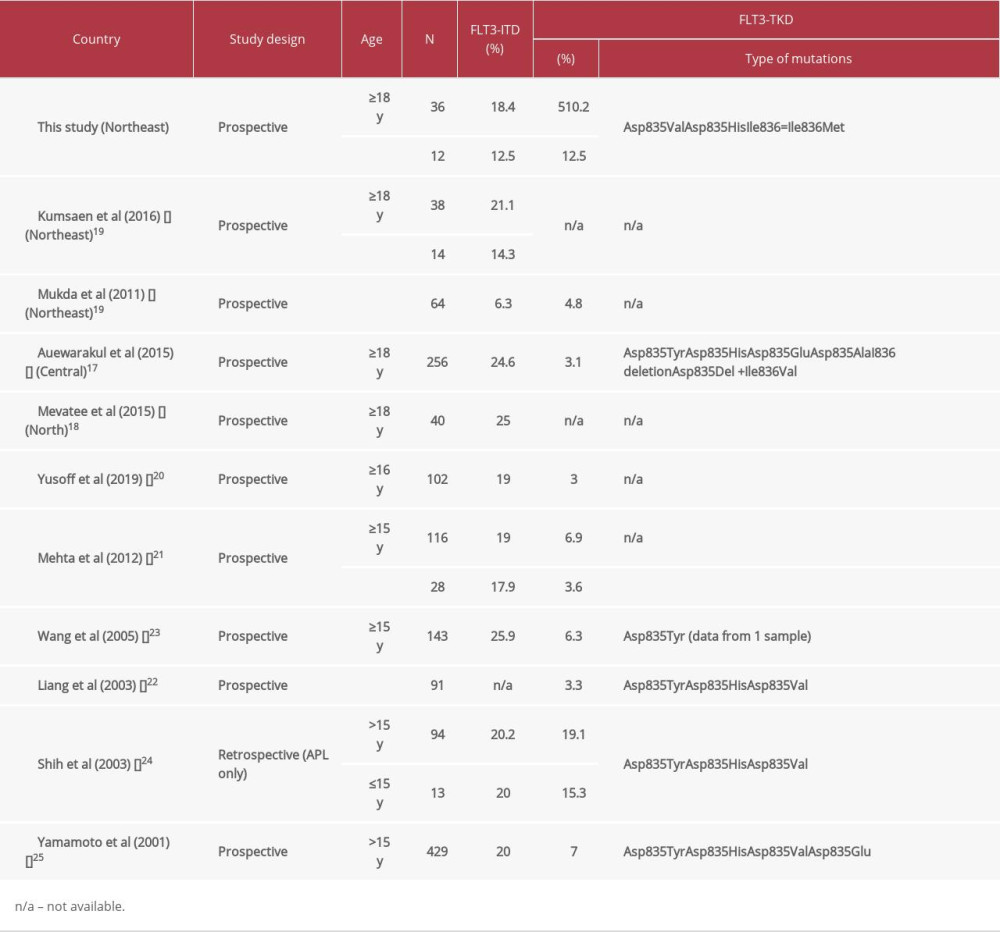
References
1. Estey EH, Acute myeloid leukemia: 2019 update on risk-stratification and management: Am J Hematol, 2018; 93(10); 1267-91
2. Grimwade D, Walker H, Oliver F, The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties: Blood, 1998; 92(7); 2322-33
3. Slovak ML, Kopecky KJ, Cassileth PA, Karyotypic analysis predicts outcome of preremission and postremission therapy in adult acute myeloid leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study: Blood, 2000; 96(13); 4075-83
4. Gregory TK, Wald D, Chen Y, Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics: J Hematol Oncol, 2009; 2; 23
5. Schnittger S, Kinkelin U, Schoch C, Screening for MLL tandem duplication in 387 unselected patients with AML identify a prognostically unfavorable subset of AML: Leukemia, 2000; 14(5); 796-804
6. Döhner K, Tobis K, Ulrich R, Prognostic significance of partial tandem duplications of the MLL gene in adult patients 16 to 60 years old with acute myeloid leukemia and normal cytogenetics: A study of the Acute Myeloid Leukemia Study Group Ulm: J Clin Oncol, 2002; 20(15); 3254-61
7. Markovic A, MacKenzie KL, Lock RB, FLT-3: A new focus in the understanding of acute leukemia: Int J Biochem Cell Biol, 2005; 37(6); 1168-72
8. Daver N, Schlenk RF, Russell NH, Levis MJ, Targeting FLT3 mutations in AML: Review of current knowledge and evidence: Leukemia, 2019; 33(2); 299-312
9. Yanada M, Matsuo K, Suzuki T, Prognostic significance of FLT3 internal tandem duplication and tyrosine kinase domain mutations for acute myeloid leukemia: A meta-analysis: Leukemia, 2005; 19(8); 1345-49
10. Fröhling S, Schlenk RF, Breitruck J, Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: A study of the AML Study Group Ulm: Blood, 2002; 100(13); 4372-80
11. Santos FP, Jones D, Qiao W, Prognostic value of FLT3 mutations among different cytogenetic subgroups in acute myeloid leukemia: Cancer, 2011; 117(10); 2145-55
12. Mead AJ, Linch DC, Hills RK, FLT3 tyrosine kinase domain mutations are biologically distinct from and have a significantly more favorable prognosis than FLT3 internal tandem duplications in patients with acute myeloid leukemia: Blood, 2007; 110(4); 1262-70
13. Chen Y, Pan Y, Guo Y, Tyrosine kinase inhibitors targeting FLT3 in the treatment of acute myeloid leukemia: Stem Cell Investig, 2017; 4; 48
14. Thiede C, Steudel C, Mohr B, Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis: Blood, 2002; 99(12); 4326-35
15. Bagrintseva K, Geisenhof S, Kern R, FLT3-ITD-TKD dual mutants associated with AML confer resistance to FLT3 PTK inhibitors and cytotoxic agents by overexpression of Bcl-x(L): Blood, 2005; 105(9); 3679-85
16. Kumsaen P, Fucharoen G, Sirijerachai C, FLT3-ITD mutations in acute myeloid leukemia patients in Northeast Thailand: Asian Pac J Cancer Prev, 2016; 17(9); 4395-99
17. Auewarakul CU, Sritana N, Limwongse C, Mutations of the FLT3 gene in adult acute myeloid leukemia: Determination of incidence and identification of a novel mutation in a Thai population: Cancer Genet Cytogenet, 2005; 162(2); 127-34
18. Mevatee P, Tantiworawit A, Traisathit P, FLT3-ITD, NPM1, and DNMT3A gene mutations and risk factors in normal karyotype acute myeloid leukemia and myelodysplastic syndrome patients in upper Northern Thailand: Asian Pac J Cancer Prev, 2017; 18(11); 3031-39
19. Mukda E, Pintaraks K, Sawangpanich R, FLT3 and NPM1 gene mutations in childhood acute myeloblastic leukemia: Asian Pac J Cancer Prev, 2011; 12(7); 1827-31
20. Mat Yusoff Y, Abu Seman Z, Othman N, Identification of FLT3 and NPM1 mutations in patients with acute myeloid leukaemia: Asian Pac J Cancer Prev, 2019; 20(6); 1749-55
21. Shalvi VM, Shilin NS, Hemangini HV, Comprehensive FLT3 analysis in indian acute myeloid leukaemia: Journal of Blood & Lymph, 2012; 2(1); 1-13
22. Liang DC, Shih LY, Hung IJ, FLT3-TKD mutation in childhood acute myeloid leukemia: Leukemia, 2003; 17(5); 883-86
23. Wang L, Lin D, Zhang X, Analysis of FLT3 internal tandem duplication and D835 mutations in Chinese acute leukemia patients: Leuk Res, 2005; 29(12); 1393-98
24. Shih LY, Kuo MC, Liang DC, Internal tandem duplication and Asp835 mutations of the FMS-like tyrosine kinase 3 (FLT3) gene in acute promyelocytic leukemia: Cancer, 2003; 98(6); 1206-16
25. Yamamoto Y, Kiyoi H, Nakano Y, Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies: Blood, 2001; 97(8); 2434-39
26. Andersson A, Johansson B, Lassen C, Clinical impact of internal tandem duplications and activating point mutations in FLT3 in acute myeloid leukemia in elderly patients: Eur J Haematol, 2004; 72(5); 307-13
27. Bacher U, Haferlach C, Kern W, Prognostic relevance of FLT3-TKD mutations in AML: the combination matters – an analysis of 3082 patients: Blood, 2008; 111(5); 2527-37
28. Kiyoi H, Naoe T, Yokota S, Internal tandem duplication of FLT3 associated with leukocytosis in acute promyelocytic leukemia. Leukemia Study Group of the Ministry of Health and Welfare (Kohseisho): Leukemia, 1997; 11(9); 1447-52
29. Tate JG, Bamford S, Jubb HC, COSMIC: The Catalogue Of Somatic Mutations In Cancer: Nucleic Acids Res, 2019; 47(D1); D941-D7
30. Kottaridis PD, Gale RE, Frew ME, The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: Analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials: Blood, 2001; 98(6); 1752-59
31. Moreno I, Martín G, Bolufer P, Incidence and prognostic value of FLT3 internal tandem duplication and D835 mutations in acute myeloid leukemia: Haematologica, 2003; 88(1); 19-24
32. Levis M, FLT3 mutations in acute myeloid leukemia: What is the best approach in 2013?: Hematology Am Soc Hematol Educ Program, 2013; 2013; 220-26
33. Grafone T, Palmisano M, Nicci C, Storti S, An overview on the role of FLT3-tyrosine kinase receptor in acute myeloid leukemia: biology and treatment: Oncol Rev, 2012; 6(1); e8
34. Stirewalt DL, Radich JP, The role of FLT3 in haematopoietic malignancies: Nat Rev Cancer, 2003; 3(9); 650-65
35. Whitman SP, Ruppert AS, Radmacher MD, FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications: Blood, 2008; 111(3); 1552-59
36. Baqai J, Crisan D, Correlation of FLT3 mutations with expression of CD7 in acute myeloid leukemia: Appl Immunohistochem Mol Morphol, 2015; 23(2); 104-8
Figures
 Figure 1. Identification of FMS-like tyrosine kinase 3 (FLT3)-internal tandem duplication (ITD) mutation by PCR. (A) Locations and orientations of primers used for identification of FLT3-ITD mutation. Primers 11F and 12R were used to amplify FLT3-ITD mutant and FLT3-wild type at exons 14–15. The 329 bp fragment indicates the wild type while the fragment larger than 329 bp indicates the FLT3-ITD mutation. TM – transmembrane domain; JM – juxtamembrane domain; TKD – tyrosine kinase domain. (B) A representative 2% agarose gel electrophoresis of PCR products. Lane 1 and lane 2 are positive and negative control for the FLT3-ITD mutation, respectively. Lanes 3–5 are PCR products of patients with acute myeloid leukemia. Lane 3 shows the 329 bp fragment indicating the wild type. Lane 4 & 5 show the 329 bp and the fragment larger than 329 bp (>329) indicating FLT3-ITD mutation. M indicates 100 bp DNA ladder.
Figure 1. Identification of FMS-like tyrosine kinase 3 (FLT3)-internal tandem duplication (ITD) mutation by PCR. (A) Locations and orientations of primers used for identification of FLT3-ITD mutation. Primers 11F and 12R were used to amplify FLT3-ITD mutant and FLT3-wild type at exons 14–15. The 329 bp fragment indicates the wild type while the fragment larger than 329 bp indicates the FLT3-ITD mutation. TM – transmembrane domain; JM – juxtamembrane domain; TKD – tyrosine kinase domain. (B) A representative 2% agarose gel electrophoresis of PCR products. Lane 1 and lane 2 are positive and negative control for the FLT3-ITD mutation, respectively. Lanes 3–5 are PCR products of patients with acute myeloid leukemia. Lane 3 shows the 329 bp fragment indicating the wild type. Lane 4 & 5 show the 329 bp and the fragment larger than 329 bp (>329) indicating FLT3-ITD mutation. M indicates 100 bp DNA ladder. Figure 2. Identification of FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutation by PCR technique followed by EcoRV digestion. (A) Locations and orientations of primers. Forward primer, F1, and reverse primer, R1, specifically to codon 20 of TKD2 domain were used to produce a 555 bp fragment. Codons 835 and 836 are encoded by nucleotides GATATC which form the EcoRV restriction site. The PCR product of wild type patients were digested by EcoRV into 2 fragments, 260 and 295 bp fragments. The acute myeloid leukemia patients with FLT3-TKD mutations, the PCR product shows 555 bp undigested fragment. TM – transmembrane domain; JM – juxtamembrane domain; TKD – tyrosine kinase domain. (B) A representative 2% agarose gel electrophoresis of PCR products among AML patients after EcoRV digestion. Lane 1 shows uncut PCR Product. Lane 2 & 3, wild type patients show PCR-RFLP fragments which the EcoRV digested produced 260 and 295 bp fragments. Lanes 4 and 5 are positive for FLT3-internal tandem duplication (ITD) mutation which show 555 bp undigested fragments. M indicates the 100 bp DNA ladder.
Figure 2. Identification of FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutation by PCR technique followed by EcoRV digestion. (A) Locations and orientations of primers. Forward primer, F1, and reverse primer, R1, specifically to codon 20 of TKD2 domain were used to produce a 555 bp fragment. Codons 835 and 836 are encoded by nucleotides GATATC which form the EcoRV restriction site. The PCR product of wild type patients were digested by EcoRV into 2 fragments, 260 and 295 bp fragments. The acute myeloid leukemia patients with FLT3-TKD mutations, the PCR product shows 555 bp undigested fragment. TM – transmembrane domain; JM – juxtamembrane domain; TKD – tyrosine kinase domain. (B) A representative 2% agarose gel electrophoresis of PCR products among AML patients after EcoRV digestion. Lane 1 shows uncut PCR Product. Lane 2 & 3, wild type patients show PCR-RFLP fragments which the EcoRV digested produced 260 and 295 bp fragments. Lanes 4 and 5 are positive for FLT3-internal tandem duplication (ITD) mutation which show 555 bp undigested fragments. M indicates the 100 bp DNA ladder. Figure 3. (A, B) Direct sequencing in the forward direction demonstrating 2 novel FMS-like tyrosine kinase 3 (FLT3)-TKD mutations among AML patients. Red arrow indicates the mutation site. The mutation and 836ATC – ATG mutation, respectively. (C) The corresponding DNA sequence of AML patients with wild type FLT3-TKD at the same region.
Figure 3. (A, B) Direct sequencing in the forward direction demonstrating 2 novel FMS-like tyrosine kinase 3 (FLT3)-TKD mutations among AML patients. Red arrow indicates the mutation site. The mutation and 836ATC – ATG mutation, respectively. (C) The corresponding DNA sequence of AML patients with wild type FLT3-TKD at the same region. Tables
 Table 1. Characteristics and laboratory data of 65 newly diagnosed acute myeloid leukemia (AML) patients according to type of FLt3 mutation.
Table 1. Characteristics and laboratory data of 65 newly diagnosed acute myeloid leukemia (AML) patients according to type of FLt3 mutation. Table 2. Characteristics of patients with FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutations.
Table 2. Characteristics of patients with FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutations. Table 3. Frequency of FMS-like tyrosine kinase 3 (FLT3)-ITD and tyrosine kinase domain (TKD) mutations in patients with acute myeloid leukemia (AML) from various studies in Asia.
Table 3. Frequency of FMS-like tyrosine kinase 3 (FLT3)-ITD and tyrosine kinase domain (TKD) mutations in patients with acute myeloid leukemia (AML) from various studies in Asia. Table 1. Characteristics and laboratory data of 65 newly diagnosed acute myeloid leukemia (AML) patients according to type of FLt3 mutation.
Table 1. Characteristics and laboratory data of 65 newly diagnosed acute myeloid leukemia (AML) patients according to type of FLt3 mutation. Table 2. Characteristics of patients with FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutations.
Table 2. Characteristics of patients with FMS-like tyrosine kinase 3 (FLT3)-tyrosine kinase domain (TKD) mutations. Table 3. Frequency of FMS-like tyrosine kinase 3 (FLT3)-ITD and tyrosine kinase domain (TKD) mutations in patients with acute myeloid leukemia (AML) from various studies in Asia.
Table 3. Frequency of FMS-like tyrosine kinase 3 (FLT3)-ITD and tyrosine kinase domain (TKD) mutations in patients with acute myeloid leukemia (AML) from various studies in Asia. Most Viewed Current Articles
15 Jun 2022 : Clinical Research
Evaluation of Apical Leakage After Root Canal Obturation with Glass Ionomer, Resin, and Zinc Oxide Eugenol ...DOI :10.12659/MSMBR.936675
Med Sci Monit Basic Res 2022; 28:e936675
07 Jul 2022 : Laboratory Research
Cytotoxicity, Apoptosis, Migration Inhibition, and Autophagy-Induced by Crude Ricin from Ricinus communis S...DOI :10.12659/MSMBR.936683
Med Sci Monit Basic Res 2022; 28:e936683
01 Jun 2022 : Laboratory Research
Comparison of Sealing Abilities Among Zinc Oxide Eugenol Root-Canal Filling Cement, Antibacterial Biocerami...DOI :10.12659/MSMBR.936319
Med Sci Monit Basic Res 2022; 28:e936319
08 Dec 2022 : Original article
Use of Estimated Glomerular Filtration Rate and Urine Albumin-to-Creatinine Ratio Based on KDIGO 2012 Guide...DOI :10.12659/MSMBR.938176
Med Sci Monit Basic Res 2022; 28:e938176








